Difference between revisions of "NusA"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 104: | Line 104: | ||
* '''Operon:''' ''[[ylxS]]-[[nusA]]-[[ylxR]]-[[ylxQ]]-[[infB]]-[[ylxP]]-[[rbfA]]'' {{PubMed|8491709}} | * '''Operon:''' ''[[ylxS]]-[[nusA]]-[[ylxR]]-[[ylxQ]]-[[infB]]-[[ylxP]]-[[rbfA]]'' {{PubMed|8491709}} | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=nusA_1732281_1733396_1 nusA] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' [[SigA]] {{PubMed|8491709}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 111: | Line 113: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 09:13, 13 April 2012
- Description: transcription termination factor of RNA polymerase
| Gene name | nusA |
| Synonyms | |
| Essential | yes PubMed |
| Product | transcription termination factor |
| Function | transcription |
| Interactions involving this protein in SubtInteract: NusA | |
| MW, pI | 41 kDa, 4.583 |
| Gene length, protein length | 1113 bp, 371 aa |
| Immediate neighbours | ylxS, ylxR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 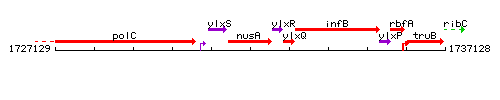
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16600
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: nusA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P32727
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Xiao Yang, Seeseei Molimau, Geoff P Doherty, Elecia B Johnston, Jon Marles-Wright, Rosalba Rothnagel, Ben Hankamer, Richard J Lewis, Peter J Lewis
The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA.
EMBO Rep: 2009, 10(9);997-1002
[PubMed:19680289]
[WorldCat.org]
[DOI]
(I p)
Karen M Davies, Amy J Dedman, Stephanie van Horck, Peter J Lewis
The NusA:RNA polymerase ratio is increased at sites of rRNA synthesis in Bacillus subtilis.
Mol Microbiol: 2005, 57(2);366-79
[PubMed:15978071]
[WorldCat.org]
[DOI]
(P p)
Alexander V Yakhnin, Paul Babitzke
NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism invitro.
Proc Natl Acad Sci U S A: 2002, 99(17);11067-72
[PubMed:12161562]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
C J Ingham, J Dennis, P A Furneaux
Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis.
Mol Microbiol: 1999, 31(2);651-63
[PubMed:10027981]
[WorldCat.org]
[DOI]
(P p)
K Shazand, J Tucker, M Grunberg-Manago, J C Rabinowitz, T Leighton
Similar organization of the nusA-infB operon in Bacillus subtilis and Escherichia coli.
J Bacteriol: 1993, 175(10);2880-7
[PubMed:8491709]
[WorldCat.org]
[DOI]
(P p)