Difference between revisions of "SdhB"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 40: | Line 40: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| − | + | {{SubtiWiki regulon|[[FsrA regulon]]}} | |
=The gene= | =The gene= | ||
| Line 114: | Line 114: | ||
* '''Regulation:''' constitutive | * '''Regulation:''' constitutive | ||
| + | ** part of the iron sparing response ([[FsrA]]) {{PubMed|22389480}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[FsrA]]: translation inhibition {{PubMed|22389480}} | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 138: | Line 140: | ||
=References= | =References= | ||
| − | <pubmed>1707123,6401289,1324713,3036777,2495271,3027051,7748886,2837411,12560550, 3910107 6799760 </pubmed> | + | <pubmed>1707123,6401289,1324713,3036777,2495271,3027051,7748886,2837411,12560550, 3910107 6799760 22389480</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:07, 14 March 2012
- Description: succinate dehydrogenase
| Gene name | sdhB |
| Synonyms | |
| Essential | no |
| Product | succinate dehydrogenase (iron-sulfur protein) |
| Function | TCA cycle |
| Interactions involving this protein in SubtInteract: SdhB | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 28 kDa, 7.989 |
| Gene length, protein length | 759 bp, 253 aa |
| Immediate neighbours | ysmA, sdhA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 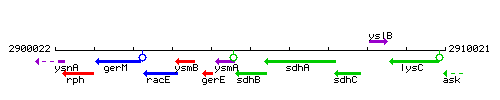
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
carbon core metabolism, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28430
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Succinate + acceptor = fumarate + reduced acceptor (according to Swiss-Prot)
- Protein family: succinate dehydrogenase/fumarate reductase iron-sulfur protein family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): Fe
- Effectors of protein activity:
- Localization: attached to the membrane PubMed
Database entries
- Structure: 1NEK (E. coli)
- UniProt: P08066
- KEGG entry: [3]
- E.C. number:EC 1.3.99.1
Additional information
- This enzyme is a trimer membrane-bound PubMed PubMed
- One subunit is bound to citochrome b558, and this subunit is the one bound to the cytosolic side of the membrane PubMed PubMed
- Another subunit is the flavoprotein one, required for FAD usage PubMed PubMed
- The other subunit has an iron-sulphur domain necessary for the catalytic activity PubMed PubMed
- extensive information on the structure and enzymatic properties of succinate dehydrogenase can be found at Proteopedia
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References