Difference between revisions of "HypR"
| Line 1: | Line 1: | ||
| − | * '''Description:''' MarR/DUF24-family transcription regulator, positively controls the nitroreductase gene ''[[ | + | * '''Description:''' MarR/DUF24-family transcription regulator, positively controls the nitroreductase gene ''[[hypO]]'' in response to disulfide stress <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | | '' | + | | ''hypR'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''yybR'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || MarR/DUF24-family transcription regulator HypR | |style="background:#ABCDEF;" align="center"| '''Product''' || MarR/DUF24-family transcription regulator HypR | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || control of the nitroreductase gene ''[[hypO]]'' in response to disulfide stress (diamide, NaOCl) |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 14 kDa, 8.415 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 14 kDa, 8.415 | ||
| Line 38: | Line 38: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| − | {{SubtiWiki regulon|[[ | + | {{SubtiWiki regulon|[[HypR regulon]]}} |
| − | = The [[ | + | = The [[HypR regulon]]:= |
| − | * ''[[ | + | * ''[[hypR]]'', ''[[hypO]]'' |
=The gene= | =The gene= | ||
| Line 58: | Line 58: | ||
=== Additional information=== | === Additional information=== | ||
| − | hypR autoregulated by disulfide stress | + | ''hypR'' is autoregulated by disulfide stress |
| Line 69: | Line 69: | ||
* '''Protein family:''' MarR/DUF24-family regulator | * '''Protein family:''' MarR/DUF24-family regulator | ||
| − | * '''Paralogous protein(s):''' YdeP,YkvN | + | * '''Paralogous protein(s):''' [[YdeP]],[[YkvN]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 75: | Line 75: | ||
* '''Kinetic information:''' Cys14 redox sensing Cys, has lower pKa of 6.36 {{PubMed|22238377}} | * '''Kinetic information:''' Cys14 redox sensing Cys, has lower pKa of 6.36 {{PubMed|22238377}} | ||
| − | * '''Domains:''' 5 alpha helices, 2 beta sheets, MarR-fold with wHTH motif, alpha4 major groove recognition helix, beta2 and 3 form the wing; alpha5 dimer interface {{PubMed|22238377}} | + | * '''Domains:''' |
| − | * '''Modification:''' oxidized to Cys14-Cys49' intersubunit disulfides by disulfide stress {{PubMed|22238377}} | + | ** 5 alpha helices, 2 beta sheets, MarR-fold with wHTH motif, alpha4 major groove recognition helix, beta2 and 3 form the wing; alpha5 dimer interface {{PubMed|22238377}} |
| + | |||
| + | * '''Modification:''' | ||
| + | ** oxidized to Cys14-Cys49' intersubunit disulfides by disulfide stress {{PubMed|22238377}} | ||
| + | ** Cys14 and Cys49' are about 8-9 Angström apart in reduced HypR-Dimer, oxidation moves the major groove recognition alpha4 helices of the HypR dimer about 4 Angstroem towards each other that leads to activation of HypR {{PubMed|22238377}} | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| Line 89: | Line 93: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | reduced HypRC14S dimer [http://www.pdb.org/pdb/explore.do?structureId=4A5N] | + | ** reduced HypRC14S dimer [http://www.pdb.org/pdb/explore.do?structureId=4A5N] {{PubMed|22238377}} |
| − | oxidized HypR C14-C49' intersubunit disulfide-linked dimer [http://www.pdb.org/pdb/explore.do?structureId=4A5M] | + | ** oxidized HypR C14-C49' intersubunit disulfide-linked dimer [http://www.pdb.org/pdb/explore.do?structureId=4A5M] {{PubMed|22238377}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P37486 P37486] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P37486 P37486] | ||
| Line 102: | Line 106: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' '' | + | * '''Operon:''' ''hypR'' (according to [http://dbtbs.hgc.jp/COG/prom/yybR.html DBTBS]) |
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
| − | * '''Regulation:''' activated by disulfide stress conditions (diamide, NaOCl) in vivo and in vitro | + | * '''Regulation:''' |
| + | ** activated by disulfide stress conditions (diamide, NaOCl) ''in vivo'' and ''in vitro '' {{PubMed|22238377}} | ||
| − | * '''Regulatory mechanism:''' redox-controlled by Cys14-Cys49' intersubunit disulfide formation by diamide and NaOCl in vitro and vivo {{PubMed|22238377}} | + | * '''Regulatory mechanism:''' |
| + | ** redox-controlled by Cys14-Cys49' intersubunit disulfide formation by diamide and NaOCl ''in vitro'' and ''in vivo'' {{PubMed|22238377}} | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 128: | Line 134: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| − | [[Haike Antelmann]],University of Greifswald, Germany | + | * [[Haike Antelmann]],University of Greifswald, Germany |
=Your additional remarks= | =Your additional remarks= | ||
| Line 138: | Line 144: | ||
=Original articles= | =Original articles= | ||
| − | <pubmed> | + | '''Additional publications:''' {{PubMed|22238377}} |
| + | <pubmed></pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:32, 16 January 2012
- Description: MarR/DUF24-family transcription regulator, positively controls the nitroreductase gene hypO in response to disulfide stress
| Gene name | hypR |
| Synonyms | yybR |
| Essential | no |
| Product | MarR/DUF24-family transcription regulator HypR |
| Function | control of the nitroreductase gene hypO in response to disulfide stress (diamide, NaOCl) |
| MW, pI | 14 kDa, 8.415 |
| Gene length, protein length | 375 bp, 125 aa |
| Immediate neighbours | cotF, ppaC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 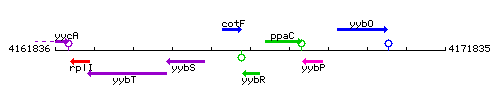
This image was kindly provided by SubtiList
| |
Contents
- 1 Categories containing this gene/protein
- 2 This gene is a member of the following regulons
- 3 This gene is a member of the following regulons
- 4 The HypR regulon:
- 5 The gene
- 6 The protein
- 7 Expression and regulation
- 8 Biological materials
- 9 Labs working on this gene/protein
- 10 Your additional remarks
- 11 References
- 12 Reviews
- 13 Original articles
Categories containing this gene/protein
transcription factors and their control, resistance against oxidative and electrophile stress
This gene is a member of the following regulons
This gene is a member of the following regulons
The HypR regulon:
The gene
Basic information
- Locus tag: BSU40540
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
hypR is autoregulated by disulfide stress
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: MarR/DUF24-family regulator
Extended information on the protein
- Kinetic information: Cys14 redox sensing Cys, has lower pKa of 6.36 PubMed
- Domains:
- 5 alpha helices, 2 beta sheets, MarR-fold with wHTH motif, alpha4 major groove recognition helix, beta2 and 3 form the wing; alpha5 dimer interface PubMed
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasmic
Database entries
- Structure:
- UniProt: P37486
- KEGG entry: [5]
- E.C. number:
Additional information
Expression and regulation
- Operon: hypR (according to DBTBS)
- Regulation:
- activated by disulfide stress conditions (diamide, NaOCl) in vivo and in vitro PubMed
- Regulatory mechanism:
- redox-controlled by Cys14-Cys49' intersubunit disulfide formation by diamide and NaOCl in vitro and in vivo PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Haike Antelmann,University of Greifswald, Germany
Your additional remarks
References
Reviews
Original articles
Additional publications: PubMed