Difference between revisions of "KatA"
(→Extended information on the protein) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' vegetative catalase 1 <br/><br/> | + | * '''Description:''' main vegetative catalase 1 <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || vegetative catalase | + | |style="background:#ABCDEF;" align="center"| '''Product''' || vegetative catalase |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || detoxification (degradation) of hydrogen peroxide | |style="background:#ABCDEF;" align="center"|'''Function''' || detoxification (degradation) of hydrogen peroxide | ||
| Line 48: | Line 48: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * increased sensitivity to oxidative stress {{PubMed|22174384}} | ||
=== Database entries === | === Database entries === | ||
| Line 56: | Line 57: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 101: | Line 100: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''katA'' | + | * '''Operon:''' ''katA'' {{PubMed|22174384}} |
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| − | ** induced in the presence of hydrogen peroxide ([[PerR]]) | + | ** induced in the presence of hydrogen peroxide and paraquat ([[PerR]]) {{PubMed|11532148,22174384}} |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | ** [[PerR]]: transcription repression | + | ** [[PerR]]: transcription repression {{PubMed|11532148}} |
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 133: | Line 132: | ||
=References= | =References= | ||
| − | <pubmed>9393707,7667267,,8931328,8932315,11532148, </pubmed> | + | <pubmed>9393707,7667267,,8931328,8932315,11532148, 22174384</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:08, 20 December 2011
- Description: main vegetative catalase 1
| Gene name | katA |
| Synonyms | kat-19 |
| Essential | no |
| Product | vegetative catalase |
| Function | detoxification (degradation) of hydrogen peroxide |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 54 kDa, 6.151 |
| Gene length, protein length | 1449 bp, 483 aa |
| Immediate neighbours | senS, ssuB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 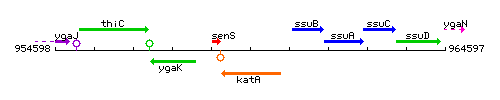
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
resistance against oxidative and electrophile stress
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU08820
Phenotypes of a mutant
- increased sensitivity to oxidative stress PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2 H2O2 = O2 + 2 H2O (according to Swiss-Prot)
- Protein family: catalase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): contains an iron-sulfur cluster
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- Structure: 1SI8 (enzyme from Enterococcus faecalis, 68% identity)
- UniProt: P26901
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: katA PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References