Difference between revisions of "MurF"
(→Categories containing this gene/protein) |
|||
| Line 92: | Line 92: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| − | * '''Localization:''' cytoplasm (according to Swiss-Prot) | + | * '''[[Localization]]:''' cytoplasm (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
| Line 113: | Line 113: | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** expression is reduced in a [[SigV]] mutant {{PubMed|21926231}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| Line 137: | Line 138: | ||
=References= | =References= | ||
| − | + | '''Additional publications:''' {{PubMed|21926231}} | |
<pubmed>18063720, 18179421, </pubmed> | <pubmed>18063720, 18179421, </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:09, 16 October 2011
- Description: UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine synthetase
| Gene name | murF |
| Synonyms | ydbQ |
| Essential | yes PubMed |
| Product | UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso- 2,6- diaminopimeloyl-D-alanyl-D-alanine synthetase |
| Function | peptidoglycan precursor biosynthesis |
| Metabolic function and regulation of this protein in SubtiPathways: Cell wall | |
| MW, pI | 49 kDa, 5.337 |
| Gene length, protein length | 1371 bp, 457 aa |
| Immediate neighbours | ddl, cshA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 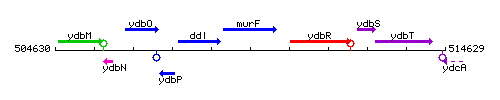
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, cell envelope stress proteins (controlled by SigM, V, W, X, Y), essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04570
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + UDP-N-acetylmuramoyl-L-alanyl-gamma-D-glutamyl-L-lysine + D-alanyl-D-alanine = ADP + phosphate + UDP-N-acetylmuramoyl-L-alanyl-gamma-D-glutamyl-L-lysyl-D-alanyl-D-alanine (according to Swiss-Prot)
- Protein family: murCDEF family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: P96613
- KEGG entry: [2]
- E.C. number: 6.3.2.10
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Jean van Heijenoort
Lipid intermediates in the biosynthesis of bacterial peptidoglycan.
Microbiol Mol Biol Rev: 2007, 71(4);620-35
[PubMed:18063720]
[WorldCat.org]
[DOI]
(P p)