Difference between revisions of "YsxC"
(→Original publications) |
(→Original publications) |
||
| Line 139: | Line 139: | ||
<pubmed> 19575570 </pubmed> | <pubmed> 19575570 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>17981968,11073929 ,12427945, 16997968 7961402 9852015 14684918 15136032 | + | '''Additional publications:''' {{PubMed|21636901}} |
| + | <pubmed>17981968,11073929 ,12427945, 16997968 7961402 9852015 14684918 15136032 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:24, 4 June 2011
- Description: assembly of the 50S subunit of the ribosome
| Gene name | ysxC |
| Synonyms | orfX |
| Essential | yes PubMed |
| Product | GTPase |
| Function | ribosome assembly |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 21 kDa, 9.708 |
| Gene length, protein length | 585 bp, 195 aa |
| Immediate neighbours | ysxD, lonA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 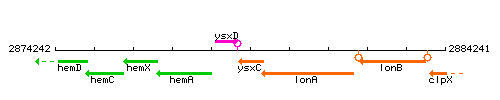
| |
Contents
Categories containing this gene/protein
translation, essential genes, membrane proteins, GTP-binding proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28190
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Binds and hydrolyzes GTP and readily exchanges GDP for GTP
- Protein family: engB family (according to Swiss-Prot) Era/Obg family
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- UniProt: P38424
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Naotake Ogasawara, Nara, Japan
Your additional remarks
References
Reviews
Robert A Britton
Role of GTPases in bacterial ribosome assembly.
Annu Rev Microbiol: 2009, 63;155-76
[PubMed:19575570]
[WorldCat.org]
[DOI]
(I p)
Original publications
Additional publications: PubMed