Difference between revisions of "FrlO"
| Line 40: | Line 40: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| − | {{SubtiWiki regulon|[[CodY regulon]]}} | + | {{SubtiWiki regulon|[[CodY regulon]]}}, {{SubtiWiki regulon|[[FrlR regulon]]}} |
=The gene= | =The gene= | ||
| Line 83: | Line 83: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' [[YurJ]]-([[FrlM]]-[[FrlN]])-[[FrlO]] {{PubMed|10092453}} | + | * '''[[Interactions]]:''' [[YurJ]]-([[FrlM]]-[[FrlN]])-[[FrlO]] {{PubMed|10092453}} |
* '''Localization:''' membrane associated (via [[FrlM]]-[[FrlN]]) {{PubMed|10092453,18763711}} | * '''Localization:''' membrane associated (via [[FrlM]]-[[FrlN]]) {{PubMed|10092453,18763711}} | ||
| Line 100: | Line 100: | ||
=Expression and regulation= | =Expression and regulation= | ||
| + | * '''Operon:''' | ||
** ''[[yurR]]-[[yurQ]]-[[frlB]]-[[frlO]]-[[frlN]]-[[frlM]]-[[frlD]]'' {{PubMed|12823818}} | ** ''[[yurR]]-[[yurQ]]-[[frlB]]-[[frlO]]-[[frlN]]-[[frlM]]-[[frlD]]'' {{PubMed|12823818}} | ||
** ''[[frlB]]-[[frlO]]-[[frlN]]-[[frlM]]-[[frlD]]'' {{PubMed|21347729}} | ** ''[[frlB]]-[[frlO]]-[[frlN]]-[[frlM]]-[[frlD]]'' {{PubMed|21347729}} | ||
| Line 106: | Line 107: | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| − | **'' [[ | + | **'' [[frlB]]'': repressed during growth in the presence of branched chain amino acids ([[CodY]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] |
** ''[[frlB]]'': induced in the presence of fructosamine ([[FrlR]]) {{PubMed|21347729}} | ** ''[[frlB]]'': induced in the presence of fructosamine ([[FrlR]]) {{PubMed|21347729}} | ||
Revision as of 18:16, 27 February 2011
- Description: aminosugar ABC transporter (binding protein)
| Gene name | frlO |
| Synonyms | yurO |
| Essential | no |
| Product | aminosugar ABC transporter (binding protein) |
| Function | uptake of sugar amines |
| Metabolic function and regulation of this protein in SubtiPathways: Alternative nitrogen sources | |
| MW, pI | 47 kDa, 5.674 |
| Gene length, protein length | 1266 bp, 422 aa |
| Immediate neighbours | frlN, frlB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 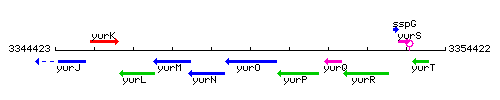
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
ABC transporters, utilization of specific carbon sources, utilization of nitrogen sources other than amino acids, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU32600
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: bacterial solute-binding protein 1 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O32156
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Veronika Maria Deppe, Johannes Bongaerts, Timothy O'Connell, Karl-Heinz Maurer, Friedhelm Meinhardt
Enzymatic deglycation of Amadori products in bacteria: mechanisms, occurrence and physiological functions.
Appl Microbiol Biotechnol: 2011, 90(2);399-406
[PubMed:21347729]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Ken-ichi Yoshida, Hirotake Yamaguchi, Masaki Kinehara, Yo-hei Ohki, Yoshiko Nakaura, Yasutaro Fujita
Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box.
Mol Microbiol: 2003, 49(1);157-65
[PubMed:12823818]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
Y Quentin, G Fichant, F Denizot
Inventory, assembly and analysis of Bacillus subtilis ABC transport systems.
J Mol Biol: 1999, 287(3);467-84
[PubMed:10092453]
[WorldCat.org]
[DOI]
(P p)