Difference between revisions of "GlcT"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || control of glucose uptake | |style="background:#ABCDEF;" align="center"|'''Function''' || control of glucose uptake | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/GlcT GlcT] | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | ||
Revision as of 12:27, 28 July 2011
- Description: transcriptional antiterminator , controls expression of the ptsG-ptsH-ptsI operon
| Gene name | glcT |
| Synonyms | ykwA |
| Essential | no |
| Product | transcriptional antiterminator of the ptsG-ptsH-ptsI operon |
| Function | control of glucose uptake |
| Interactions involving this protein in SubtInteract: GlcT | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 33,0 kDa, 7.01 |
| Gene length, protein length | 855 bp, 285 amino acids |
| Immediate neighbours | ykvZ, ptsG |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 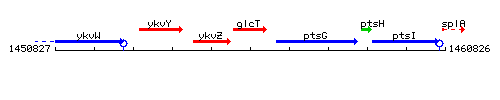
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
carbon core metabolism, transcription factors and their control, RNA binding regulators, phosphoproteins
This gene is a member of the following regulons
The GlcT regulon: ptsG-ptsH-ptsI
The gene
Basic information
- Locus tag: BSU13880
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription antiterminator , RNA-binding protein, binds the ptsG RAT sequence
- Protein family: transcription antiterminator of the BglG/ SacY family
Extended information on the protein
- Kinetic information:
- Domains:
- RNA-binding domain (N-terminal, constitutive antiterminator)
- 2x PTS regulation domains (PRDs) (C-terminal, neg. regulated by PtsG)
- Modification: phosphorylation (His104)
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- UniProt: O31691
- KEGG entry: [2]
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: available in Stülke lab:
- Expression vector:
- pGP124 (full length, in pWH844), available in Stülke lab
- pGP114 (amino acids 1-60, RNA-binding domain, in pWH844), available in Stülke lab
- pGP230 (amino acids 1-60, RNA-binding domain with thrombin cleavage site, in pWH844), available in Stülke lab
- pGP164 (both PRDs, in pWH844), in addition diverse expression vectors for phosphorylation site mutants and for RBD mutants (all in pWH844), available in Stülke lab
- pGP424 (PRDI, in pWH844), available in Stülke lab
- pGP425 (PRDII, in pWH844), available in Stülke lab
- pGP442 (PRDI, in pGP570, with thrombin cleavage site), available in Stülke lab
- pGP443 (PRDII, in pGP570, with thrombin cleavage site), available in Stülke lab
- pGP575 (amino acids 1-60, RNA-binding domain with Strep-tag, in pGP574), available in Stülke lab
- lacZ fusion:
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Reviews
Original publications
Dayté D Rodríguez, Christian Grosse, Sebastian Himmel, César González, Iñaki M de Ilarduya, Stefan Becker, George M Sheldrick, Isabel Usón
Crystallographic ab initio protein structure solution below atomic resolution.
Nat Methods: 2009, 6(9);651-3
[PubMed:19684596]
[WorldCat.org]
[DOI]
(I p)
Oliver Schilling, Christina Herzberg, Tina Hertrich, Hanna Vörsmann, Dirk Jessen, Sebastian Hübner, Fritz Titgemeyer, Jörg Stülke
Keeping signals straight in transcription regulation: specificity determinants for the interaction of a family of conserved bacterial RNA-protein couples.
Nucleic Acids Res: 2006, 34(21);6102-15
[PubMed:17074746]
[WorldCat.org]
[DOI]
(I p)
Oliver Schilling, Ines Langbein, Michael Müller, Matthias H Schmalisch, Jörg Stülke
A protein-dependent riboswitch controlling ptsGHI operon expression in Bacillus subtilis: RNA structure rather than sequence provides interaction specificity.
Nucleic Acids Res: 2004, 32(9);2853-64
[PubMed:15155854]
[WorldCat.org]
[DOI]
(I e)
Matthias H Schmalisch, Steffi Bachem, Jörg Stülke
Control of the Bacillus subtilis antiterminator protein GlcT by phosphorylation. Elucidation of the phosphorylation chain leading to inactivation of GlcT.
J Biol Chem: 2003, 278(51);51108-15
[PubMed:14527945]
[WorldCat.org]
[DOI]
(P p)
David B Greenberg, Jorg Stülke, Milton H Saier
Domain analysis of transcriptional regulators bearing PTS regulatory domains.
Res Microbiol: 2002, 153(8);519-26
[PubMed:12437213]
[WorldCat.org]
[DOI]
(P p)
I Langbein, S Bachem, J Stülke
Specific interaction of the RNA-binding domain of the bacillus subtilis transcriptional antiterminator GlcT with its RNA target, RAT.
J Mol Biol: 1999, 293(4);795-805
[PubMed:10543968]
[WorldCat.org]
[DOI]
(P p)
S Bachem, J Stülke
Regulation of the Bacillus subtilis GlcT antiterminator protein by components of the phosphotransferase system.
J Bacteriol: 1998, 180(20);5319-26
[PubMed:9765562]
[WorldCat.org]
[DOI]
(P p)
J Stülke, I Martin-Verstraete, M Zagorec, M Rose, A Klier, G Rapoport
Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT.
Mol Microbiol: 1997, 25(1);65-78
[PubMed:11902727]
[WorldCat.org]
[DOI]
(P p)
Complete list of publications: PubMed