Difference between revisions of "LytE"
(→References) |
|||
| Line 38: | Line 38: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
{{SubtiWiki regulon|[[SigH regulon]]}}, | {{SubtiWiki regulon|[[SigH regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[SigI regulon]]}}, | ||
{{SubtiWiki regulon|[[Spo0A regulon]]}}, | {{SubtiWiki regulon|[[Spo0A regulon]]}}, | ||
{{SubtiWiki regulon|[[WalR regulon]]}} | {{SubtiWiki regulon|[[WalR regulon]]}} | ||
| Line 48: | Line 49: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | a ''[[cwlO]] [[lytE]]'' mutant is not viable {{PubMed|17581128}} | + | ** a ''[[cwlO]] [[lytE]]'' mutant is not viable {{PubMed|17581128}} |
| + | ** growth defect at high temperature {{PubMed|21541672}} | ||
=== Database entries === | === Database entries === | ||
| Line 57: | Line 59: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 103: | Line 103: | ||
* '''Operon:''' ''lytE'' {{PubMed|9573210}} | * '''Operon:''' ''lytE'' {{PubMed|9573210}} | ||
| − | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|9573210}}, [[SigH]] {{PubMed|9573210}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|9573210}}, [[SigH]] {{PubMed|9573210}}, [[SigI]] {{PubMed|21541672}} |
* '''Regulation:''' | * '''Regulation:''' | ||
** repressed under conditions that trigger sporulation ([[Spo0A]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647 PubMed] | ** repressed under conditions that trigger sporulation ([[Spo0A]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647 PubMed] | ||
| − | + | ** induced at high temperature ([[SigI]]) {{PubMed|21541672}} | |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
** [[WalR]]: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/17581128 PubMed] | ** [[WalR]]: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/17581128 PubMed] | ||
| Line 134: | Line 134: | ||
=References= | =References= | ||
'''Additional publications:''' {{PubMed|21261835}} | '''Additional publications:''' {{PubMed|21261835}} | ||
| − | <pubmed>16950129,17581128,14594841,9457885,9573210,10322020, 20059685,14651647,</pubmed> | + | <pubmed>16950129,17581128,14594841,9457885,9573210,10322020, 20059685,14651647, 21541672 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:51, 5 May 2011
- Description: cell wall hydrolase (major autolysin),endopeptidase-type autolysin
| Gene name | lytE |
| Synonyms | papQ, cwlF |
| Essential | no |
| Product | cell wall hydrolase (major autolysin),endopeptidase-type autolysin |
| Function | major autolysin, cell separation, cell lysis |
| MW, pI | 37 kDa, 10.713 |
| Gene length, protein length | 1029 bp, 343 aa |
| Immediate neighbours | phoA, citR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 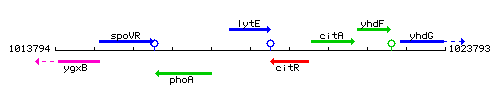
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover
This gene is a member of the following regulons
SigH regulon, SigI regulon, Spo0A regulon, WalR regulon
The gene
Basic information
- Locus tag: BSU09420
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: nlpC/p60 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: secreted (according to Swiss-Prot), binds the cell wall PubMed
Database entries
- Structure:
- UniProt: P54421
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: lytE PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed