Difference between revisions of "RapA"
| Line 32: | Line 32: | ||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| + | |||
| + | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[protein modification]]}}, | ||
| + | {{SubtiWiki category|[[transcription factors and their control]]}}, | ||
| + | {{SubtiWiki category|[[phosphorelay]]}}, | ||
| + | {{SubtiWiki category|[[quorum sensing]]}} | ||
| + | |||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[CodY regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[ComA regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[Spo0A regulon]]}} | ||
=The gene= | =The gene= | ||
| Line 51: | Line 62: | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
Revision as of 18:43, 8 December 2010
- Description: response regulator aspartate phosphatase, dephosphorylates Spo0F-P, control of the phosphorelay
| Gene name | rapA |
| Synonyms | gsiAA, spo0L |
| Essential | no |
| Product | response regulator aspartate phosphatase |
| Function | control of sporulation initiation |
| Function and regulation of this protein in SubtiPathways: Phosphorelay | |
| MW, pI | 44 kDa, 4.848 |
| Gene length, protein length | 1134 bp, 378 aa |
| Immediate neighbours | yjoB, phrA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 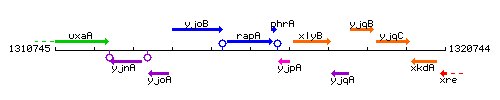
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
protein modification, transcription factors and their control, phosphorelay, quorum sensing
This gene is a member of the following regulons
CodY regulon, ComA regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU12430
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: RAP family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- UniProt: Q00828
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ilka B Bischofs, Joshua A Hug, Aiwen W Liu, Denise M Wolf, Adam P Arkin
Complexity in bacterial cell-cell communication: quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay.
Proc Natl Acad Sci U S A: 2009, 106(16);6459-64
[PubMed:19380751]
[WorldCat.org]
[DOI]
(I p)
Natalia Comella, Alan D Grossman
Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis.
Mol Microbiol: 2005, 57(4);1159-74
[PubMed:16091051]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
Shu Ishikawa, Leighton Core, Marta Perego
Biochemical characterization of aspartyl phosphate phosphatase interaction with a phosphorylated response regulator and its inhibition by a pentapeptide.
J Biol Chem: 2002, 277(23);20483-9
[PubMed:11923303]
[WorldCat.org]
[DOI]
(P p)
M Perego, P Glaser, J A Hoch
Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis.
Mol Microbiol: 1996, 19(6);1151-7
[PubMed:8730857]
[WorldCat.org]
[DOI]
(P p)
M Perego, J A Hoch
Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1996, 93(4);1549-53
[PubMed:8643670]
[WorldCat.org]
[DOI]
(P p)
J P Mueller, G Bukusoglu, A L Sonenshein
Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system.
J Bacteriol: 1992, 174(13);4361-73
[PubMed:1378051]
[WorldCat.org]
[DOI]
(P p)