Difference between revisions of "PerR"
| Line 32: | Line 32: | ||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| + | |||
| + | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[transcription factors and their control]]}}, | ||
| + | {{SubtiWiki category|[[resistance against oxidative and electrophile stress]]}} | ||
| + | |||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[PerR regulon]]}} | ||
=The gene= | =The gene= | ||
| Line 51: | Line 58: | ||
| − | + | ||
| − | |||
| − | |||
=The protein= | =The protein= | ||
Revision as of 17:52, 8 December 2010
- Description: transcriptional repressor of the peroxide regulon
| Gene name | perR |
| Synonyms | ygaG |
| Essential | no |
| Product | transcriptional repressor |
| Function | regulation of the response to peroxide |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 16 kDa, 5.888 |
| Gene length, protein length | 435 bp, 145 aa |
| Immediate neighbours | ygaF, ygzB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 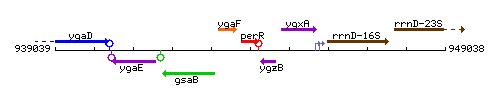
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription factors and their control, resistance against oxidative and electrophile stress
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU08730
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: Fur family (according to Swiss-Prot)
Genes repressed by PerR
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: selective metal catalyzed oxidation of two histidine residues of the regulatory site results in induction (loss of DNA-binding activity) PubMed
- Cofactor(s): contains an iron-sulfur cluster
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: P71086
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: perR PubMed
- Regulation:
- Additional information:
Biological materials
- Mutant: HB0509 (spc), available in John Helmann's and Jörg Stülke's labs, also GP868 (fur::mls, perR::spc).
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Your additional remarks
References
Reviews
Melinda J Faulkner, John D Helmann
Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis.
Antioxid Redox Signal: 2011, 15(1);175-89
[PubMed:20977351]
[WorldCat.org]
[DOI]
(I p)
Victor Duarte, Jean-Marc Latour
PerR vs OhrR: selective peroxide sensing in Bacillus subtilis.
Mol Biosyst: 2010, 6(2);316-23
[PubMed:20094649]
[WorldCat.org]
[DOI]
(I p)
Peter Zuber
Management of oxidative stress in Bacillus.
Annu Rev Microbiol: 2009, 63;575-97
[PubMed:19575568]
[WorldCat.org]
[DOI]
(I p)
David P Giedroc
Hydrogen peroxide sensing in Bacillus subtilis: it is all about the (metallo)regulator.
Mol Microbiol: 2009, 73(1);1-4
[PubMed:19508286]
[WorldCat.org]
[DOI]
(I p)
Original Publications
L Jacquamet, D A K Traoré, J-L Ferrer, O Proux, D Testemale, J-L Hazemann, E Nazarenko, A El Ghazouani, C Caux-Thang, V Duarte, J-M Latour
Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding.
Mol Microbiol: 2009, 73(1);20-31
[PubMed:19508285]
[WorldCat.org]
[DOI]
(I p)
Falko Hochgräfe, Carmen Wolf, Stephan Fuchs, Manuel Liebeke, Michael Lalk, Susanne Engelmann, Michael Hecker
Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus.
J Bacteriol: 2008, 190(14);4997-5008
[PubMed:18487332]
[WorldCat.org]
[DOI]
(I p)
Montira Leelakriangsak, Kazuo Kobayashi, Peter Zuber
Dual negative control of spx transcription initiation from the P3 promoter by repressors PerR and YodB in Bacillus subtilis.
J Bacteriol: 2007, 189(5);1736-44
[PubMed:17158660]
[WorldCat.org]
[DOI]
(P p)
Jin-Won Lee, John D Helmann
Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR.
J Biol Chem: 2006, 281(33);23567-78
[PubMed:16766519]
[WorldCat.org]
[DOI]
(P p)
Jin-Won Lee, John D Helmann
The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation.
Nature: 2006, 440(7082);363-7
[PubMed:16541078]
[WorldCat.org]
[DOI]
(I p)
Kentaro Hayashi, Taku Ohsawa, Kazuo Kobayashi, Naotake Ogasawara, Mitsuo Ogura
The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis.
J Bacteriol: 2005, 187(19);6659-67
[PubMed:16166527]
[WorldCat.org]
[DOI]
(P p)
Charles M Moore, Michiko M Nakano, Tao Wang, Rick W Ye, John D Helmann
Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside.
J Bacteriol: 2004, 186(14);4655-64
[PubMed:15231799]
[WorldCat.org]
[DOI]
(P p)
Mayuree Fuangthong, John D Helmann
Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis.
J Bacteriol: 2003, 185(21);6348-57
[PubMed:14563870]
[WorldCat.org]
[DOI]
(P p)
John D Helmann, Ming Fang Winston Wu, Ahmed Gaballa, Phil A Kobel, Maud M Morshedi, Paul Fawcett, Chris Paddon
The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors.
J Bacteriol: 2003, 185(1);243-53
[PubMed:12486061]
[WorldCat.org]
[DOI]
(P p)
Ahmed Gaballa, John D Helmann
A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis.
Mol Microbiol: 2002, 45(4);997-1005
[PubMed:12180919]
[WorldCat.org]
[DOI]
(P p)
Mayuree Fuangthong, Andrew F Herbig, Nada Bsat, John D Helmann
Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible.
J Bacteriol: 2002, 184(12);3276-86
[PubMed:12029044]
[WorldCat.org]
[DOI]
(P p)
A F Herbig, J D Helmann
Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA.
Mol Microbiol: 2001, 41(4);849-59
[PubMed:11532148]
[WorldCat.org]
[DOI]
(P p)
L Casillas-Martinez, A Driks, B Setlow, P Setlow
Lack of a significant role for the PerR regulator in Bacillus subtilis spore resistance.
FEMS Microbiol Lett: 2000, 188(2);203-8
[PubMed:10913706]
[WorldCat.org]
[DOI]
(P p)
N Bsat, A Herbig, L Casillas-Martinez, P Setlow, J D Helmann
Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors.
Mol Microbiol: 1998, 29(1);189-98
[PubMed:9701813]
[WorldCat.org]
[DOI]
(P p)
N Bsat, L Chen, J D Helmann
Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes.
J Bacteriol: 1996, 178(22);6579-86
[PubMed:8932315]
[WorldCat.org]
[DOI]
(P p)