Difference between revisions of "CtsR"
| Line 32: | Line 32: | ||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| + | |||
| + | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[proteolysis]]}}, | ||
| + | {{SubtiWiki category|[[transcription factors and their control]]}}, | ||
| + | {{SubtiWiki category|[[general stress proteins (controlled by SigB)]]}}, | ||
| + | {{SubtiWiki category|[[heat shock proteins]]}}, | ||
| + | {{SubtiWiki category|[[phosphoproteins]]}} | ||
| + | |||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[CtsR regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[SigB regulon]]}} | ||
=The gene= | =The gene= | ||
| Line 51: | Line 62: | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
Revision as of 15:20, 8 December 2010
| Gene name | ctsR |
| Synonyms | yacG |
| Essential | no |
| Product | transcription repressor |
| Function | regulation of protein degradation |
| Regulatory function and regulation of this protein in SubtiPathways: Stress, Phosphorelay | |
| MW, pI | 17 kDa, 9.261 |
| Gene length, protein length | 462 bp, 154 aa |
| Immediate neighbours | rrnW-5S, mcsA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 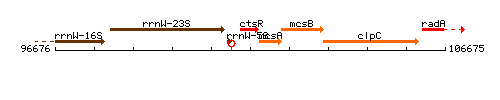
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
proteolysis, transcription factors and their control, general stress proteins (controlled by SigB), heat shock proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00830
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ctsR family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- UniProt: P37568
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information: the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Alexander K W Elsholz, Stephan Michalik, Daniela Zühlke, Michael Hecker, Ulf Gerth
CtsR, the Gram-positive master regulator of protein quality control, feels the heat.
EMBO J: 2010, 29(21);3621-9
[PubMed:20852588]
[WorldCat.org]
[DOI]
(I p)
Marcus Miethke, Michael Hecker, Ulf Gerth
Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control.
J Bacteriol: 2006, 188(13);4610-9
[PubMed:16788169]
[WorldCat.org]
[DOI]
(P p)
Pekka Varmanen, Finn K Vogensen, Karin Hammer, Airi Palva, Hanne Ingmer
ClpE from Lactococcus lactis promotes repression of CtsR-dependent gene expression.
J Bacteriol: 2003, 185(17);5117-24
[PubMed:12923084]
[WorldCat.org]
[DOI]
(P p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)
J D Helmann, M F Wu, P A Kobel, F J Gamo, M Wilson, M M Morshedi, M Navre, C Paddon
Global transcriptional response of Bacillus subtilis to heat shock.
J Bacteriol: 2001, 183(24);7318-28
[PubMed:11717291]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
E Krüger, D Zühlke, E Witt, H Ludwig, M Hecker
Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor.
EMBO J: 2001, 20(4);852-63
[PubMed:11179229]
[WorldCat.org]
[DOI]
(P p)
I Derré, G Rapoport, T Msadek
The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C.
Mol Microbiol: 2000, 38(2);335-47
[PubMed:11069659]
[WorldCat.org]
[DOI]
(P p)
I Derré, G Rapoport, T Msadek
CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria.
Mol Microbiol: 1999, 31(1);117-31
[PubMed:9987115]
[WorldCat.org]
[DOI]
(P p)
E Krüger, M Hecker
The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes.
J Bacteriol: 1998, 180(24);6681-8
[PubMed:9852015]
[WorldCat.org]
[DOI]
(P p)
E Krüger, T Msadek, M Hecker
Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon.
Mol Microbiol: 1996, 20(4);713-23
[PubMed:8793870]
[WorldCat.org]
[DOI]
(P p)
E Krüger, U Völker, M Hecker
Stress induction of clpC in Bacillus subtilis and its involvement in stress tolerance.
J Bacteriol: 1994, 176(11);3360-7
[PubMed:8195092]
[WorldCat.org]
[DOI]
(P p)
Additional publications: PubMed