Difference between revisions of "KsgA"
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || rRNA adenine dimethyltransferase | |style="background:#ABCDEF;" align="center"| '''Product''' || rRNA adenine dimethyltransferase | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || rRNA maturation |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 32 kDa, 5.712 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 32 kDa, 5.712 | ||
Revision as of 15:15, 18 June 2010
- Description: rRNA adenine dimethyltransferase
| Gene name | ksgA |
| Synonyms | |
| Essential | no |
| Product | rRNA adenine dimethyltransferase |
| Function | rRNA maturation |
| MW, pI | 32 kDa, 5.712 |
| Gene length, protein length | 876 bp, 292 aa |
| Immediate neighbours | rnmV, yabG |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 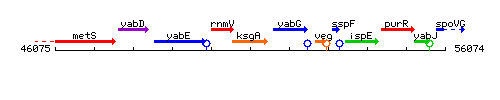
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU00420
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: methylation of A1518 and A1519 in 16S rRNA
- Protein family: KsgA subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P37468
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Kozo Ochi, Ji-Yun Kim, Yukinori Tanaka, Guojun Wang, Kenta Masuda, Hideaki Nanamiya, Susumu Okamoto, Shinji Tokuyama, Yoshikazu Adachi, Fujio Kawamura
Inactivation of KsgA, a 16S rRNA methyltransferase, causes vigorous emergence of mutants with high-level kasugamycin resistance.
Antimicrob Agents Chemother: 2009, 53(1);193-201
[PubMed:19001112]
[WorldCat.org]
[DOI]
(I p)
Stephanie T Wang, Barbara Setlow, Erin M Conlon, Jessica L Lyon, Daisuke Imamura, Tsutomu Sato, Peter Setlow, Richard Losick, Patrick Eichenberger
The forespore line of gene expression in Bacillus subtilis.
J Mol Biol: 2006, 358(1);16-37
[PubMed:16497325]
[WorldCat.org]
[DOI]
(P p)
C Condon, D Brechemier-Baey, B Beltchev, M Grunberg-Manago, H Putzer
Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: mature 5S rRNA is dispensable for ribosome function.
RNA: 2001, 7(2);242-53
[PubMed:11233981]
[WorldCat.org]
[DOI]
(P p)