Difference between revisions of "LeuA"
(→Database entries) |
|||
| Line 95: | Line 95: | ||
* '''[[Sigma factor]]:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/1577690 PubMed] | * '''[[Sigma factor]]:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/1577690 PubMed] | ||
| − | * '''Regulation:''' | + | * '''Regulation:''' for a complete overview on the regulation of the ilv operon, see [http://www.ncbi.nlm.nih.gov/pubmed/20935095 Brinsmade ''et al''.] |
| + | ** repressed by casamino acids [http://www.ncbi.nlm.nih.gov/pubmed/12107147 PubMed] | ||
| + | ** expression is stimulated in the presence of glucose [http://www.ncbi.nlm.nih.gov/sites/entrez/12193635 PubMed] | ||
** repressed in the absence of good nitrogen sources (glutamine or ammonium) ([[TnrA]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/15547269 PubMed] | ** repressed in the absence of good nitrogen sources (glutamine or ammonium) ([[TnrA]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/15547269 PubMed] | ||
| − | ** repressed during growth in the presence of branched chain amino acids ([[CodY]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | + | ** repressed during growth in the presence of branched chain amino acids ([[CodY]], [[T-box]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] |
| − | * '''Regulatory mechanism:''' [[CodY]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | + | * '''Regulatory mechanism:''' |
| + | ** [[CodY]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | ||
| + | ** [[CcpA]]: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/12193635 PubMed] | ||
| + | ** [[T-box]]: tRNA-controlled [[RNA switch]] that mediates termination/antitermination | ||
** [[TnrA]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/15547269 PubMed] | ** [[TnrA]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/15547269 PubMed] | ||
| − | |||
* '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] | * '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] | ||
| Line 125: | Line 129: | ||
=References= | =References= | ||
| − | <pubmed>15060025,12193635,19258532,8289305,18641142,15547269,12618455,,15547269,12618455,12107147 18763711, </pubmed> | + | <pubmed>15060025,12193635,19258532,8289305,18641142,15547269,12618455,,15547269,12618455,12107147 18763711, 20935095</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:36, 26 October 2010
- Description: 2-isopropylmalate synthase
| Gene name | leuA |
| Synonyms | |
| Essential | no |
| Product | 2-isopropylmalate synthase |
| Function | biosynthesis of leucine |
| Metabolic function and regulation of this protein in SubtiPathways: Ile, Leu, Val, Coenzyme A | |
| MW, pI | 56 kDa, 5.657 |
| Gene length, protein length | 1554 bp, 518 aa |
| Immediate neighbours | leuB, ilvC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 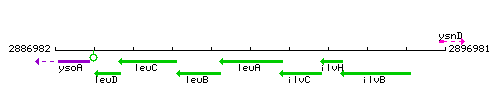
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU28280
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Acetyl-CoA + 3-methyl-2-oxobutanoate + H2O = (2S)-2-isopropylmalate + CoA (according to Swiss-Prot)
- Protein family: LeuA type 1 subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot), membrane PubMed
Database entries
- Structure: 3EEG (from Cytophaga hutchinsonii atcc 33406, 53% identity, 68% similarity)
- UniProt: P94565
- KEGG entry: [3]
- E.C. number: 2.3.3.13
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation: for a complete overview on the regulation of the ilv operon, see Brinsmade et al.
- Regulatory mechanism:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References