Difference between revisions of "YxnB"
(→Expression and regulation) |
|||
| Line 89: | Line 89: | ||
=Expression and regulation= | =Expression and regulation= | ||
| + | * '''Operon:''' | ||
| + | ** ''[[yxbB]]-[[yxbA]]-[[yxnB]]-[[asnH]]-[[yxaM]]'' {{PubMed|20185509,10746760}} | ||
| + | ** ''[[yxbB]]-[[yxbA]]-[[yxnB]]'' {{PubMed|20185509}} | ||
| − | + | * '''[[Sigma factor]]:''' SigA {{PubMed|20185509}} | |
| − | |||
| − | * '''[[Sigma factor]]:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** repressed by glucose (8-fold) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed] | ||
** repressed during growth in the presence of branched chain amino acids ([[CodY]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | ** repressed during growth in the presence of branched chain amino acids ([[CodY]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | ||
** repressed during logrithmic growth ([[AbrB]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/15101989 PubMed] | ** repressed during logrithmic growth ([[AbrB]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/15101989 PubMed] | ||
| Line 103: | Line 105: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** the tricistronic ''[[yxbB]]-[[yxbA]]-[[yxnB]]'' transcript is extremely stable, and stability is mediated by a 5'-proximal long sequence triplication preceding the ''[[yxbB]]'' open reading frame {{PubMed|20185509}} | ||
=Biological materials = | =Biological materials = | ||
| Line 124: | Line 127: | ||
=References= | =References= | ||
| − | <pubmed>10746760,12618455,10498721,,12618455,15101989, </pubmed> | + | <pubmed>10746760,12618455,10498721,20185509,12618455,15101989, </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:11, 27 February 2010
- Description: unknown
| Gene name | yxnB |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| Metabolic function and regulation of this protein in SubtiPathways: Asp, Asn | |
| MW, pI | 18 kDa, 5.109 |
| Gene length, protein length | 480 bp, 160 aa |
| Immediate neighbours | yxbA, asnH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 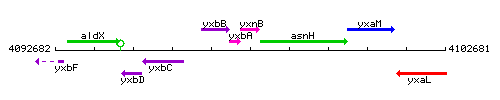
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU39910
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O34704
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor: SigA PubMed
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Tetsuro Morinaga, Kazuo Kobayashi, Hitoshi Ashida, Yasutaro Fujita, Ken-Ichi Yoshida
Transcriptional regulation of the Bacillus subtilis asnH operon and role of the 5'-proximal long sequence triplication in RNA stabilization.
Microbiology (Reading): 2010, 156(Pt 6);1632-1641
[PubMed:20185509]
[WorldCat.org]
[DOI]
(I p)
Mélanie A Hamon, Nicola R Stanley, Robert A Britton, Alan D Grossman, Beth A Lazazzera
Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis.
Mol Microbiol: 2004, 52(3);847-60
[PubMed:15101989]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
Ken-Ichi Yoshida, Izumi Ishio, Eishi Nagakawa, Yoshiyuki Yamamoto, Mami Yamamoto, Yasutaro Fujita
Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome.
Microbiology (Reading): 2000, 146 ( Pt 3);573-579
[PubMed:10746760]
[WorldCat.org]
[DOI]
(P p)
K Yoshida, Y Fujita, S D Ehrlich
Three asparagine synthetase genes of Bacillus subtilis.
J Bacteriol: 1999, 181(19);6081-91
[PubMed:10498721]
[WorldCat.org]
[DOI]
(P p)