Difference between revisions of "PlsY"
(→Basic information/ Evolution) |
(→Extended information on the protein) |
||
| Line 72: | Line 72: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''Interactions:''' [[PlsY]]-[[PlsX]] {{PubMed|19282621}} |
| − | * '''Localization:''' cell membrane {{PubMed|19820159}} | + | * '''Localization:''' cell membrane {{PubMed|19820159}} |
=== Database entries === | === Database entries === | ||
Revision as of 13:48, 14 January 2010
- Description: acylphosphate:glycerol-phosphate acyltransferase
| Gene name | plsY |
| Synonyms | yneS |
| Essential | yes PubMed |
| Product | acylphosphate:glycerol-phosphate acyltransferase |
| Function | biosynthesis of phospholipids |
| MW, pI | 20 kDa, 10.408 |
| Gene length, protein length | 579 bp, 193 aa |
| Immediate neighbours | yneR, yneT |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 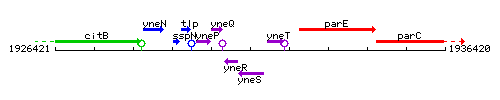
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU18070
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Acyl-phosphate + sn-glycerol 3-phosphate = acyl-sn-glycerol 3-phosphate + phosphate (according to UniProt)
- Protein family: UPF0078 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane PubMed
Database entries
- Structure:
- UniProt: Q45064
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Matsumoto lab PubMed
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Yasutaro Fujita, Hiroshi Matsuoka, Kazutake Hirooka
Regulation of fatty acid metabolism in bacteria.
Mol Microbiol: 2007, 66(4);829-39
[PubMed:17919287]
[WorldCat.org]
[DOI]
(P p)
Stephen W White, Jie Zheng, Yong-Mei Zhang, Rock
The structural biology of type II fatty acid biosynthesis.
Annu Rev Biochem: 2005, 74;791-831
[PubMed:15952903]
[WorldCat.org]
[DOI]
(P p)
Original Publications