Difference between revisions of "YwrD"
| Line 85: | Line 85: | ||
=== Additional information=== | === Additional information=== | ||
| + | The gene is annotated in KEGG as an ortholog of gamma-glutamyltranspeptidase EC 2.3.2.2, This enzyme is necessary to utilize glutathione (GSH) as the sulfur source. No EC annotation for this gene is available in Swiss-Prot/MetaCyc. It has been shown by Minami et al. ({{PubMed|19935659}}) that ywrD mutant grows well on minimal media supplied with GSH as the sole sulfur source. In addition, His-tag purified ywrD cannot hydrolyze GSH. {{PubMed|19935659}} | ||
| + | |||
=Expression and regulation= | =Expression and regulation= | ||
Revision as of 12:31, 26 November 2009
- Description: unknown
| Gene name | ywrD |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| MW, pI | 57 kDa, 5.451 |
| Gene length, protein length | 1575 bp, 525 aa |
| Immediate neighbours | ywrE, ywrC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 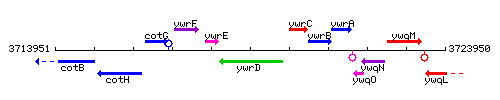
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU36100
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (5-L-glutamyl)-peptide + an amino acid = peptide + 5-L-glutamyl amino acid (according to Swiss-Prot), however, all evidence suggests this is a misannotation PubMed
- Protein family: gamma-glutamyltransferase family (according to Swiss-Prot), , however, all evidence suggests this is a misannotation PubMed
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O05218
- KEGG entry: [3]
- E.C. number: 2.3.2.2
Additional information
The gene is annotated in KEGG as an ortholog of gamma-glutamyltranspeptidase EC 2.3.2.2, This enzyme is necessary to utilize glutathione (GSH) as the sulfur source. No EC annotation for this gene is available in Swiss-Prot/MetaCyc. It has been shown by Minami et al. (PubMed) that ywrD mutant grows well on minimal media supplied with GSH as the sole sulfur source. In addition, His-tag purified ywrD cannot hydrolyze GSH. PubMed
Expression and regulation
- Operon:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References