Difference between revisions of "OdhA"
| Line 20: | Line 20: | ||
|style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 2823 bp, 941 aa | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 2823 bp, 941 aa | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[odhB]], [[yojO]]'' |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB13829]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB13829]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' | ||
Revision as of 17:21, 4 December 2009
- Description: 2-oxoglutarate dehydrogenase (E1 subunit)
| Gene name | odhA |
| Synonyms | citK |
| Essential | no |
| Product | 2-oxoglutarate dehydrogenase (E1 subunit) |
| Function | TCA cycle |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 105 kDa, 5.871 |
| Gene length, protein length | 2823 bp, 941 aa |
| Immediate neighbours | odhB, yojO |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 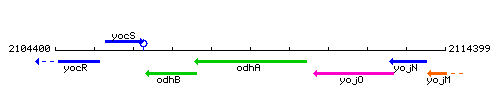
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU19370
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-oxoglutarate + [dihydrolipoyllysine-residue succinyltransferase] lipoyllysine = [dihydrolipoyllysine-residue succinyltransferase] S-succinyldihydrolipoyllysine + CO2 (according to Swiss-Prot)
- Protein family: 2-oxoacid dehydrogenase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- UniProt: P23129
- KEGG entry: [3]
- E.C. number: 1.2.4.2
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant: GP671 (spc), GP665 (aphA3), GP684 (cat), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Ciarán Condon, Jordi Rourera, Dominique Brechemier-Baey, Harald Putzer
Ribonuclease M5 has few, if any, mRNA substrates in Bacillus subtilis.
J Bacteriol: 2002, 184(10);2845-9
[PubMed:11976317]
[WorldCat.org]
[DOI]
(P p)
O Resnekov, L Melin, P Carlsson, M Mannerlöv, A von Gabain, L Hederstedt
Organization and regulation of the Bacillus subtilis odhAB operon, which encodes two of the subenzymes of the 2-oxoglutarate dehydrogenase complex.
Mol Gen Genet: 1992, 234(2);285-96
[PubMed:1508153]
[WorldCat.org]
[DOI]
(P p)
P Carlsson, L Hederstedt
Genetic characterization of Bacillus subtilis odhA and odhB, encoding 2-oxoglutarate dehydrogenase and dihydrolipoamide transsuccinylase, respectively.
J Bacteriol: 1989, 171(7);3667-72
[PubMed:2500417]
[WorldCat.org]
[DOI]
(P p)