Difference between revisions of "ClpY"
m |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' two-component ATP-dependent protease <br/><br/> | + | * '''Description:''' two-component ATP-dependent protease, ATPase subunit <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || two-component ATP-dependent protease | + | |style="background:#ABCDEF;" align="center"| '''Product''' || two-component ATP-dependent protease, <br/>ATPase subunit |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || protein degradation | |style="background:#ABCDEF;" align="center"|'''Function''' || protein degradation | ||
| Line 70: | Line 70: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''Interactions:''' [[ClpQ]]-[[ClpY]] {{PubMed|18689473}} |
* '''Localization:''' | * '''Localization:''' | ||
Revision as of 06:02, 15 January 2010
- Description: two-component ATP-dependent protease, ATPase subunit
| Gene name | clpY |
| Synonyms | hslU, codX |
| Essential | no |
| Product | two-component ATP-dependent protease, ATPase subunit |
| Function | protein degradation |
| MW, pI | 52 kDa, 5.31 |
| Gene length, protein length | 1401 bp, 467 aa |
| Immediate neighbours | clpQ, codY |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 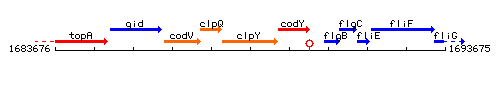
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU16160
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: HslU subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- UniProt: P39778
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Min Suk Kang, Soon Rae Kim, Pyeongsu Kwack, Byung Kook Lim, Sung Won Ahn, Young Min Rho, Ihn Sik Seong, Seong-Chul Park, Soo Hyun Eom, Gang-Won Cheong, Chin Ha Chung
Molecular architecture of the ATP-dependent CodWX protease having an N-terminal serine active site.
EMBO J: 2003, 22(12);2893-902
[PubMed:12805205]
[WorldCat.org]
[DOI]
(P p)
M Ratnayake-Lecamwasam, P Serror, K W Wong, A L Sonenshein
Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels.
Genes Dev: 2001, 15(9);1093-103
[PubMed:11331605]
[WorldCat.org]
[DOI]
(P p)
M S Kang, B K Lim, I S Seong, J H Seol, N Tanahashi, K Tanaka, C H Chung
The ATP-dependent CodWX (HslVU) protease in Bacillus subtilis is an N-terminal serine protease.
EMBO J: 2001, 20(4);734-42
[PubMed:11179218]
[WorldCat.org]
[DOI]
(P p)
F J Slack, P Serror, E Joyce, A L Sonenshein
A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon.
Mol Microbiol: 1995, 15(4);689-702
[PubMed:7783641]
[WorldCat.org]
[DOI]
(P p)