Difference between revisions of "StoA"
(→Expression and regulation) |
(→Database entries) |
||
| Line 76: | Line 76: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=3ERW 3ERW] |
* '''UniProt:''' [http://www.uniprot.org/uniprot/O31687 O31687] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O31687 O31687] | ||
Revision as of 20:14, 13 October 2009
- Description: thiol-disulfide oxidoreductase, with thioredoxin-like domain, required for the synthesis of the endospore peptidoglycan cortex
| Gene name | stoA |
| Synonyms | spoIVH, ykvV |
| Essential | no |
| Product | thiol-disulfide oxidoreductase |
| Function | spore cortex formation |
| MW, pI | 18 kDa, 5.297 |
| Gene length, protein length | 495 bp, 165 aa |
| Immediate neighbours | ykvU, zosA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 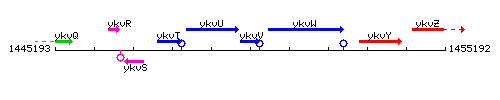
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU13840
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: thioredoxin domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: spore wall (according to Swiss-Prot)
Database entries
- Structure: 3ERW
- UniProt: O31687
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Allister Crow, Yiming Liu, Mirja Carlsson Möller, Nick E Le Brun, Lars Hederstedt
Structure and functional properties of Bacillus subtilis endospore biogenesis factor StoA.
J Biol Chem: 2009, 284(15);10056-66
[PubMed:19144642]
[WorldCat.org]
[DOI]
(P p)
Lyethur S Erlendsson, Mirja Möller, Lars Hederstedt
Bacillus subtilis StoA Is a thiol-disulfide oxidoreductase important for spore cortex synthesis.
J Bacteriol: 2004, 186(18);6230-8
[PubMed:15342593]
[WorldCat.org]
[DOI]
(P p)
Daisuke Imamura, Kazuo Kobayashi, Junichi Sekiguchi, Naotake Ogasawara, Michio Takeuchi, Tsutomu Sato
spoIVH (ykvV), a requisite cortex formation gene, is expressed in both sporulating compartments of Bacillus subtilis.
J Bacteriol: 2004, 186(16);5450-9
[PubMed:15292147]
[WorldCat.org]
[DOI]
(P p)
Patrick Eichenberger, Shane T Jensen, Erin M Conlon, Christiaan van Ooij, Jessica Silvaggi, José Eduardo González-Pastor, Masaya Fujita, Sigal Ben-Yehuda, Patrick Stragier, Jun S Liu, Richard Losick
The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis.
J Mol Biol: 2003, 327(5);945-72
[PubMed:12662922]
[WorldCat.org]
[DOI]
(P p)