Difference between revisions of "FapR"
(→Expression and regulation) |
(→Expression and regulation) |
||
| Line 87: | Line 87: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | + | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigA]] |
| − | + | ||
* '''Regulation:''' | * '''Regulation:''' | ||
** repressed unless the cells experience an accumulation of malonyl-CoA ([[FapR]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12737802 PubMed] | ** repressed unless the cells experience an accumulation of malonyl-CoA ([[FapR]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12737802 PubMed] | ||
Revision as of 06:26, 3 July 2009
- Description: repressor of fatty acid synthetic genes
| Gene name | fapR |
| Synonyms | ylpC |
| Essential | no |
| Product | transcriptional repressor |
| Function | regulation of fatty acid biosynthesis |
| MW, pI | 21 kDa, 5.393 |
| Gene length, protein length | 564 bp, 188 aa |
| Immediate neighbours | recG, plsX |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 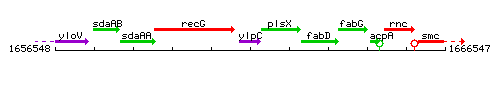
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU15880
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: fapR family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: malonyl-CoA, acts as the molecular inducer of the FapR regulon
- Interactions:
- Localization:
Database entries
- Swiss prot entry: O34835
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Yong-Mei Zhang, Charles O Rock
Transcriptional regulation in bacterial membrane lipid synthesis.
J Lipid Res: 2009, 50 Suppl(Suppl);S115-9
[PubMed:18941141]
[WorldCat.org]
[DOI]
(P p)
Letal I Salzberg, John D Helmann
Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition.
J Bacteriol: 2008, 190(23);7797-807
[PubMed:18820022]
[WorldCat.org]
[DOI]
(I p)
Gustavo E Schujman, Silvia Altabe, Diego de Mendoza
A malonyl-CoA-dependent switch in the bacterial response to a dysfunction of lipid metabolism.
Mol Microbiol: 2008, 68(4);987-96
[PubMed:18384517]
[WorldCat.org]
[DOI]
(I p)
Yasutaro Fujita, Hiroshi Matsuoka, Kazutake Hirooka
Regulation of fatty acid metabolism in bacteria.
Mol Microbiol: 2007, 66(4);829-39
[PubMed:17919287]
[WorldCat.org]
[DOI]
(P p)
Gustavo E Schujman, Marcelo Guerin, Alejandro Buschiazzo, Francis Schaeffer, Leticia I Llarrull, Georgina Reh, Alejandro J Vila, Pedro M Alzari, Diego de Mendoza
Structural basis of lipid biosynthesis regulation in Gram-positive bacteria.
EMBO J: 2006, 25(17);4074-83
[PubMed:16932747]
[WorldCat.org]
[DOI]
(P p)
Gustavo E Schujman, Luciana Paoletti, Alan D Grossman, Diego de Mendoza
FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis.
Dev Cell: 2003, 4(5);663-72
[PubMed:12737802]
[WorldCat.org]
[DOI]
(P p)