Difference between revisions of "SpoVAD"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || spore maturation | |style="background:#ABCDEF;" align="center"|'''Function''' || spore maturation | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/lys_threo.html Lys, Thr]''' | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 35 kDa, 5.243 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 35 kDa, 5.243 | ||
Revision as of 15:13, 17 June 2009
- Description: required for spore maturation
| Gene name | spoVAD |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | spore maturation |
| Metabolic function and regulation of this protein in SubtiPathways: Lys, Thr | |
| MW, pI | 35 kDa, 5.243 |
| Gene length, protein length | 1014 bp, 338 aa |
| Immediate neighbours | spoVAE, spoVAC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 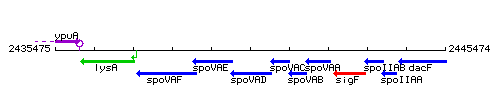
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU23410
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: P40869
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Venkata Ramana Vepachedu, Peter Setlow
Analysis of interactions between nutrient germinant receptors and SpoVA proteins of Bacillus subtilis spores.
FEMS Microbiol Lett: 2007, 274(1);42-7
[PubMed:17573930]
[WorldCat.org]
[DOI]
(P p)
Venkata Ramana Vepachedu, Peter Setlow
Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme.
J Bacteriol: 2007, 189(5);1565-72
[PubMed:17158659]
[WorldCat.org]
[DOI]
(P p)
Venkata Ramana Vepachedu, Peter Setlow
Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis.
J Bacteriol: 2005, 187(16);5677-82
[PubMed:16077113]
[WorldCat.org]
[DOI]
(P p)
Venkata Ramana Vepachedu, Peter Setlow
Analysis of the germination of spores of Bacillus subtilis with temperature sensitive spo mutations in the spoVA operon.
FEMS Microbiol Lett: 2004, 239(1);71-7
[PubMed:15451103]
[WorldCat.org]
[DOI]
(P p)
Federico Tovar-Rojo, Monica Chander, Barbara Setlow, Peter Setlow
The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis.
J Bacteriol: 2002, 184(2);584-7
[PubMed:11751839]
[WorldCat.org]
[DOI]
(P p)
J Errington, J Mandelstam
Use of a lacZ gene fusion to determine the dependence pattern and the spore compartment expression of sporulation operon spoVA in spo mutants of Bacillus subtilis.
J Gen Microbiol: 1986, 132(11);2977-85
[PubMed:3114420]
[WorldCat.org]
[DOI]
(P p)
P Fort, J Errington
Nucleotide sequence and complementation analysis of a polycistronic sporulation operon, spoVA, in Bacillus subtilis.
J Gen Microbiol: 1985, 131(5);1091-105
[PubMed:3926949]
[WorldCat.org]
[DOI]
(P p)
J Errington, J Mandelstam
Genetic and phenotypic characterization of a cluster of mutations in the spoVA locus of Bacillus subtilis.
J Gen Microbiol: 1984, 130(8);2115-21
[PubMed:6432957]
[WorldCat.org]
[DOI]
(P p)