Difference between revisions of "YtiA"
| Line 120: | Line 120: | ||
=References= | =References= | ||
| − | <pubmed>15805528, </pubmed> | + | <pubmed>12904577,15049826,15805528, </pubmed> |
# Höper et al. (2005) Comprehensive Characterization of the Contribution of Individual [[SigB]]-Dependent General Stress Genes to Stress Resistance of ''Bacillus subtilis''. J. Bact. '''187:''' 2810-2826 [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] | # Höper et al. (2005) Comprehensive Characterization of the Contribution of Individual [[SigB]]-Dependent General Stress Genes to Stress Resistance of ''Bacillus subtilis''. J. Bact. '''187:''' 2810-2826 [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 15:06, 12 June 2009
- Description: general stress protein, binds in the stationary phase to the ribosome, replaces RpmE under conditions of zinc limitation
| Gene name | ytiA |
| Synonyms | |
| Essential | no |
| Product | accessory ribosomal protein |
| Function | survival of salt stress |
| MW, pI | 9 kDa, 9.808 |
| Gene length, protein length | 246 bp, 82 aa |
| Immediate neighbours | ytiB, ythA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 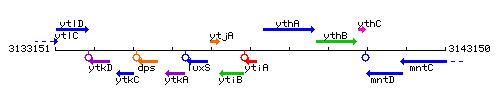
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU30700
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: Type B subfamily (according to Swiss-Prot)
- Paralogous protein(s): RpmE
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry: O34967
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Hideaki Nanamiya, Genki Akanuma, Yousuke Natori, Rikinori Murayama, Saori Kosono, Toshiaki Kudo, Kazuo Kobayashi, Naotake Ogasawara, Seung-Moon Park, Kozo Ochi, Fujio Kawamura
Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome.
Mol Microbiol: 2004, 52(1);273-83
[PubMed:15049826]
[WorldCat.org]
[DOI]
(P p)
Ekaterina M Panina, Andrey A Mironov, Mikhail S Gelfand
Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins.
Proc Natl Acad Sci U S A: 2003, 100(17);9912-7
[PubMed:12904577]
[WorldCat.org]
[DOI]
(P p)