Difference between revisions of "TrnO-Ile"
| Line 81: | Line 81: | ||
=References= | =References= | ||
| + | <pubmed>6312418, </pubmed> | ||
# Ogasawara et al. (1983) Structure and organization of rRNA operons in the region of the replication origin of the ''Bacillus subtilis'' chromosome.''Nucl. Acids Res.'' '''11:''' 6301-6318. [http://www.ncbi.nlm.nih.gov/sites/entrez/6312418 PubMed] | # Ogasawara et al. (1983) Structure and organization of rRNA operons in the region of the replication origin of the ''Bacillus subtilis'' chromosome.''Nucl. Acids Res.'' '''11:''' 6301-6318. [http://www.ncbi.nlm.nih.gov/sites/entrez/6312418 PubMed] | ||
Revision as of 21:21, 8 June 2009
- Description: tRNA-Ile
| Gene name | trnO-Ile |
| Synonyms | |
| Essential | no |
| Product | transfer RNA-Ile |
| Function | translation |
| Gene length | 77 bp |
| Immediate neighbours | rrnO-16S, trnO-Ala |
| Gene sequence (+200bp) | |
Genetic context 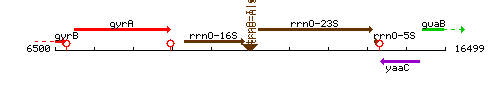
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag:
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
- Structure:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
N Ogasawara, S Moriya, H Yoshikawa
Structure and organization of rRNA operons in the region of the replication origin of the Bacillus subtilis chromosome.
Nucleic Acids Res: 1983, 11(18);6301-18
[PubMed:6312418]
[WorldCat.org]
[DOI]
(P p)
- Ogasawara et al. (1983) Structure and organization of rRNA operons in the region of the replication origin of the Bacillus subtilis chromosome.Nucl. Acids Res. 11: 6301-6318. PubMed