Difference between revisions of "XynB"
| Line 120: | Line 120: | ||
=References= | =References= | ||
| + | <pubmed>8012596, </pubmed> | ||
# Lindner, C., Stülke, J. & Hecker, M. (1994) Regulation of xylanolytic enzymes in Bacillus subtilis. Microbiology 140: 753-757. [http://www.ncbi.nlm.nih.gov/sites/entrez/8012596 PubMed] | # Lindner, C., Stülke, J. & Hecker, M. (1994) Regulation of xylanolytic enzymes in Bacillus subtilis. Microbiology 140: 753-757. [http://www.ncbi.nlm.nih.gov/sites/entrez/8012596 PubMed] | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 19:36, 8 June 2009
- Description: xylan beta-1,4-xylosidase
| Gene name | xynB |
| Synonyms | ynaK |
| Essential | no |
| Product | xylan beta-1,4-xylosidase |
| Function | xylan degradation |
| MW, pI | 61 kDa, 5.326 |
| Gene length, protein length | 1599 bp, 533 aa |
| Immediate neighbours | xynP, xylR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 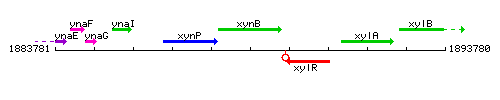
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU17580
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolysis of (1->4)-beta-D-xylans, to remove successive D-xylose residues from the non-reducing termini (according to Swiss-Prot)
- Protein family: glycosyl hydrolase 43 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure: 1YIF
- Swiss prot entry: P94489
- KEGG entry: [3]
- E.C. number: 3.2.1.37
Additional information
Expression and regulation
- Regulation: repressed by glucose (CcpA) , carbon catabolite repression (CcpA), induction by xylose (XylR)
- Regulatory mechanism: CcpA: transcription repression, CcpA: transcription repression, XylR: transcription repression
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
C Lindner, J Stülke, M Hecker
Regulation of xylanolytic enzymes in Bacillus subtilis.
Microbiology (Reading): 1994, 140 ( Pt 4);753-7
[PubMed:8012596]
[WorldCat.org]
[DOI]
(P p)