Difference between revisions of "DegS"
(→References) |
|||
| Line 124: | Line 124: | ||
=References= | =References= | ||
| − | + | <pubmed>16479537, 17218307 </pubmed> | |
| − | + | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 20:17, 1 June 2009
- Description: two-component sensor kinase for exoenzyme and competence regulation
| Gene name | degS |
| Synonyms | sacU |
| Essential | no |
| Product | two-component sensor kinase (NarL family) |
| Function | regulation of degradative enzyme and genetic competence |
| MW, pI | 44 kDa, 5.988 |
| Gene length, protein length | 1155 bp, 385 aa |
| Immediate neighbours | degU, yvyE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 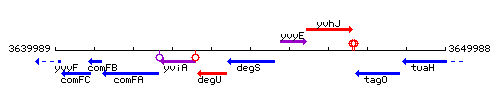
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + protein L-histidine = ADP + protein N-phospho-L-histidine (according to Swiss-Prot) autophosphorylation, autodephosphorylation, phosphorylation of DegU on Asp-56 PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- aa 12 ... 170: unknown DegS domain
- aa 180 ... 248: HisKA_3 domain (autophosphorylation)
- aa 289 ... 384: HATPase_c
- Modification: phosphorylation on Ser-76 PubMed, autophosphorylation on His-189
- Cofactor(s):
- Effectors of protein activity: DegQ
- Localization: Cytoplasm (Homogeneous) PubMed
Database entries
- Structure:
- Swiss prot entry: P13799
- KEGG entry: BSU35500
- E.C. number:
Additional information
Expression and regulation
- Regulation: induced in the presence of ammonium (TnrA)
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Tarek Msadek, Institut Pasteur, Paris, France
Your additional remarks
References
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed