Difference between revisions of "ParB"
| Line 4: | Line 4: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''parB'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''spo0J, spo0JB '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
| Line 20: | Line 20: | ||
(antagonist of [[Soj]]-dependent inhibition of sporulation initiation) | (antagonist of [[Soj]]-dependent inhibition of sporulation initiation) | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU40960 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU40960 parB] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=Spo0J Spo0J] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=Spo0J Spo0J] | ||
| Line 28: | Line 28: | ||
|style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 846 bp, 282 aa | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 846 bp, 282 aa | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yyaC]]'', ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yyaC]]'', ''[[parA]]'' |
|- | |- | ||
|style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU40960 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU40960 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU40960 DNA_with_flanks] | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU40960 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU40960 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU40960 DNA_with_flanks] | ||
| Line 74: | Line 74: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | ** inhibits [[ | + | ** inhibits [[ParA]] dimerization and concomitant DNA-binding activity by stimulating [[ParA]] ATPase activity {{PubMed|21235642}} |
** recruits the condensin complex to the DNA origin region {{PubMed|24440393}} | ** recruits the condensin complex to the DNA origin region {{PubMed|24440393}} | ||
** the dimer binds specifcally to the centromere-like ''parS'' sequence {{PubMed|25572315}} | ** the dimer binds specifcally to the centromere-like ''parS'' sequence {{PubMed|25572315}} | ||
| Line 96: | Line 96: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
** forms dimers {{PubMed|24829297}} | ** forms dimers {{PubMed|24829297}} | ||
| − | ** [[ | + | ** [[ParA]]-[[ParB]] {{PubMed|21235642}} |
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| Line 117: | Line 117: | ||
* '''Operon:''' ''[[soj]]-[[spo0J]]'' {{PubMed|22383849}} | * '''Operon:''' ''[[soj]]-[[spo0J]]'' {{PubMed|22383849}} | ||
| − | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=parB_4205556_4206404_-1 | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=parB_4205556_4206404_-1 parB] {{PubMed|22383849}} |
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
| Line 131: | Line 131: | ||
** number of protein molecules per cell (complex medium with amino acids, without glucose): 362 {{PubMed|24696501}} | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 362 {{PubMed|24696501}} | ||
** number of protein molecules per cell: 880 (equivalent to 440 dimers) {{PubMed|24829297}} | ** number of protein molecules per cell: 880 (equivalent to 440 dimers) {{PubMed|24829297}} | ||
| + | |||
=Biological materials = | =Biological materials = | ||
* '''Mutant:''' | * '''Mutant:''' | ||
Revision as of 08:03, 10 August 2015
- Description: chromosome positioning near the pole and transport through the polar septum / antagonist of Soj-dependent inhibition of sporulation initiation, traps DNA loops, required for the recruitment of the SMC complex to the origin of replication
| Gene name | parB |
| Synonyms | spo0J, spo0JB |
| Essential | no |
| Product | site-specific DNA-binding protein |
| Function | chromosome positioning before asymmetric septation,
specifying its orientation, and imposing directionality on its subsequent transport through the septum; required for sporulation (antagonist of Soj-dependent inhibition of sporulation initiation) |
| Gene expression levels in SubtiExpress: parB | |
| Interactions involving this protein in SubtInteract: Spo0J | |
| MW, pI | 32 kDa, 8.417 |
| Gene length, protein length | 846 bp, 282 aa |
| Immediate neighbours | yyaC, parA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed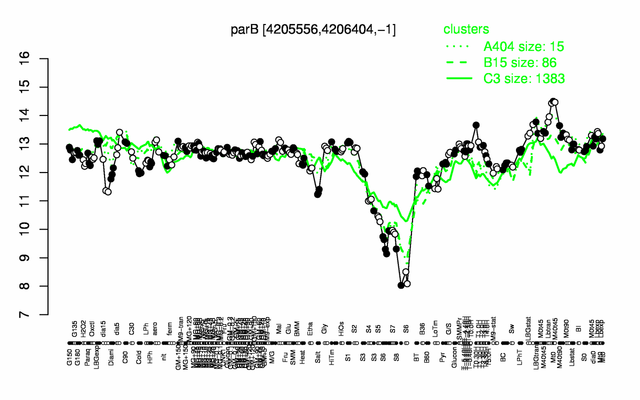
| |
Contents
Categories containing this gene/protein
DNA condensation/ segregation, sporulation/ other
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40960
Phenotypes of a mutant
Database entries
- BsubCyc: BSU40960
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: parB family (according to Swiss-Prot)
- Paralogous protein(s): Noc
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- chromosome centromer PubMed
Database entries
- BsubCyc: BSU40960
- Structure:
- UniProt: P26497
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Heath Murray, Centre for Bacterial Cell Biology, Newcastle, UK homepage
Your additional remarks
References
Reviews
Rodrigo Reyes-Lamothe, Emilien Nicolas, David J Sherratt
Chromosome replication and segregation in bacteria.
Annu Rev Genet: 2012, 46;121-43
[PubMed:22934648]
[WorldCat.org]
[DOI]
(I p)
Original publications
Larissa Wilhelm, Frank Bürmann, Anita Minnen, Ho-Chul Shin, Christopher P Toseland, Byung-Ha Oh, Stephan Gruber
SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis.
Elife: 2015, 4;
[PubMed:25951515]
[WorldCat.org]
[DOI]
(I e)
James A Taylor, Cesar L Pastrana, Annika Butterer, Christian Pernstich, Emma J Gwynn, Frank Sobott, Fernando Moreno-Herrero, Mark S Dillingham
Specific and non-specific interactions of ParB with DNA: implications for chromosome segregation.
Nucleic Acids Res: 2015, 43(2);719-31
[PubMed:25572315]
[WorldCat.org]
[DOI]
(I p)
Xindan Wang, Paula Montero Llopis, David Z Rudner
Bacillus subtilis chromosome organization oscillates between two distinct patterns.
Proc Natl Acad Sci U S A: 2014, 111(35);12877-82
[PubMed:25071173]
[WorldCat.org]
[DOI]
(I p)
Thomas G W Graham, Xindan Wang, Dan Song, Candice M Etson, Antoine M van Oijen, David Z Rudner, Joseph J Loparo
ParB spreading requires DNA bridging.
Genes Dev: 2014, 28(11);1228-38
[PubMed:24829297]
[WorldCat.org]
[DOI]
(I p)
Jan Muntel, Vincent Fromion, Anne Goelzer, Sandra Maaβ, Ulrike Mäder, Knut Büttner, Michael Hecker, Dörte Becher
Comprehensive absolute quantification of the cytosolic proteome of Bacillus subtilis by data independent, parallel fragmentation in liquid chromatography/mass spectrometry (LC/MS(E)).
Mol Cell Proteomics: 2014, 13(4);1008-19
[PubMed:24696501]
[WorldCat.org]
[DOI]
(I p)
Xindan Wang, Olive W Tang, Eammon P Riley, David Z Rudner
The SMC condensin complex is required for origin segregation in Bacillus subtilis.
Curr Biol: 2014, 24(3);287-92
[PubMed:24440393]
[WorldCat.org]
[DOI]
(I p)
Luise A K Kleine Borgmann, Hanna Hummel, Maximilian H Ulbrich, Peter L Graumann
SMC condensation centers in Bacillus subtilis are dynamic structures.
J Bacteriol: 2013, 195(10);2136-45
[PubMed:23475963]
[WorldCat.org]
[DOI]
(I p)
Hajime Okumura, Mika Yoshimura, Mikako Ueki, Taku Oshima, Naotake Ogasawara, Shu Ishikawa
Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis.
Nucleic Acids Res: 2012, 40(1);220-34
[PubMed:21911367]
[WorldCat.org]
[DOI]
(I p)
Graham Scholefield, Rachel Whiting, Jeff Errington, Heath Murray
Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation.
Mol Microbiol: 2011, 79(4);1089-100
[PubMed:21235642]
[WorldCat.org]
[DOI]
(I p)
Nora L Sullivan, Kathleen A Marquis, David Z Rudner
Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation.
Cell: 2009, 137(4);697-707
[PubMed:19450517]
[WorldCat.org]
[DOI]
(I p)
Stephan Gruber, Jeff Errington
Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis.
Cell: 2009, 137(4);685-96
[PubMed:19450516]
[WorldCat.org]
[DOI]
(I p)
Heath Murray, Jeff Errington
Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA.
Cell: 2008, 135(1);74-84
[PubMed:18854156]
[WorldCat.org]
[DOI]
(I p)
Shu Ishikawa, Yoshitoshi Ogura, Mika Yoshimura, Hajime Okumura, Eunha Cho, Yoshikazu Kawai, Ken Kurokawa, Taku Oshima, Naotake Ogasawara
Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method.
DNA Res: 2007, 14(4);155-68
[PubMed:17932079]
[WorldCat.org]
[DOI]
(P p)
Adam M Breier, Alan D Grossman
Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome.
Mol Microbiol: 2007, 64(3);703-18
[PubMed:17462018]
[WorldCat.org]
[DOI]
(P p)
Heath Murray, Henrique Ferreira, Jeff Errington
The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites.
Mol Microbiol: 2006, 61(5);1352-61
[PubMed:16925562]
[WorldCat.org]
[DOI]
(P p)
Philina S Lee, Alan D Grossman
The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis.
Mol Microbiol: 2006, 60(4);853-69
[PubMed:16677298]
[WorldCat.org]
[DOI]
(P p)
Gonçalo Real, Sabine Autret, Elizabeth J Harry, Jeffery Errington, Adriano O Henriques
Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis.
Mol Microbiol: 2005, 55(2);349-67
[PubMed:15659156]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Yoshitoshi Ogura, Naotake Ogasawara, Elizabeth J Harry, Shigeki Moriya
Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis.
J Bacteriol: 2003, 185(21);6316-24
[PubMed:14563866]
[WorldCat.org]
[DOI]
(P p)
Ling Juan Wu, Jeff Errington
RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis.
Mol Microbiol: 2003, 49(6);1463-75
[PubMed:12950914]
[WorldCat.org]
[DOI]
(P p)
Philina S Lee, Daniel Chi-Hong Lin, Shigeki Moriya, Alan D Grossman
Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis.
J Bacteriol: 2003, 185(4);1326-37
[PubMed:12562803]
[WorldCat.org]
[DOI]
(P p)
S Autret, R Nair, J Errington
Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein.
Mol Microbiol: 2001, 41(3);743-55
[PubMed:11532141]
[WorldCat.org]
[DOI]
(P p)
J D Quisel, A D Grossman
Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB).
J Bacteriol: 2000, 182(12);3446-51
[PubMed:10852876]
[WorldCat.org]
[DOI]
(P p)
R A Britton, A D Grossman
Synthetic lethal phenotypes caused by mutations affecting chromosome partitioning in Bacillus subtilis.
J Bacteriol: 1999, 181(18);5860-4
[PubMed:10482533]
[WorldCat.org]
[DOI]
(P p)
A A Teleman, P L Graumann, D C Lin, A D Grossman, R Losick
Chromosome arrangement within a bacterium.
Curr Biol: 1998, 8(20);1102-9
[PubMed:9778525]
[WorldCat.org]
[DOI]
(P p)
M A Cervin, G B Spiegelman, B Raether, K Ohlsen, M Perego, J A Hoch
A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis.
Mol Microbiol: 1998, 29(1);85-95
[PubMed:9701805]
[WorldCat.org]
[DOI]
(P p)
C D Webb, P L Graumann, J A Kahana, A A Teleman, P A Silver, R Losick
Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis.
Mol Microbiol: 1998, 28(5);883-92
[PubMed:9663676]
[WorldCat.org]
[DOI]
(P p)
D C Lin, A D Grossman
Identification and characterization of a bacterial chromosome partitioning site.
Cell: 1998, 92(5);675-85
[PubMed:9506522]
[WorldCat.org]
[DOI]
(P p)
P J Lewis, J Errington
Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein.
Mol Microbiol: 1997, 25(5);945-54
[PubMed:9364919]
[WorldCat.org]
[DOI]
(P p)
P Glaser, M E Sharpe, B Raether, M Perego, K Ohlsen, J Errington
Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning.
Genes Dev: 1997, 11(9);1160-8
[PubMed:9159397]
[WorldCat.org]
[DOI]
(P p)
M E Sharpe, J Errington
The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning.
Mol Microbiol: 1996, 21(3);501-9
[PubMed:8866474]
[WorldCat.org]
[DOI]
(P p)
K Ireton, N W Gunther, A D Grossman
spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis.
J Bacteriol: 1994, 176(17);5320-9
[PubMed:8071208]
[WorldCat.org]
[DOI]
(P p)
T H Mysliwiec, J Errington, A B Vaidya, M G Bramucci
The Bacillus subtilis spo0J gene: evidence for involvement in catabolite repression of sporulation.
J Bacteriol: 1991, 173(6);1911-9
[PubMed:1900505]
[WorldCat.org]
[DOI]
(P p)