Difference between revisions of "Mfd"
(→Original publications) |
|||
| Line 158: | Line 158: | ||
<pubmed>17239578 15063847 17917874 8811173 7968917 15720542 16464004 19023283 22201950 22749141 22933559 </pubmed> | <pubmed>17239578 15063847 17917874 8811173 7968917 15720542 16464004 19023283 22201950 22749141 22933559 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>8594198,20435731 ,104170,16950921,9535092, 16469698 21699902 22178973 22248542 22960746 21710567,20702425 24296669 21145481 24118570 25326311 25713353</pubmed> | + | <pubmed>8594198,20435731 ,104170,16950921,9535092, 16469698 21699902 22178973 22248542 22960746 21710567,20702425 24296669 21145481 24118570 25326311 25961799 25713353</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:41, 11 June 2015
- Description: transcription-repair coupling factor, eliminates genetic damage from transcriptionally active genes during sporulation, required for increased mutagenesis of lagging strand genes
| Gene name | mfd |
| Synonyms | |
| Essential | no |
| Product | transcription-repair coupling factor |
| Function | promotes strand-specific DNA repair by displacing
RNA polymerase stalled at a nucleotide lesion and directing the (A)BC excinuclease to the RNA damage site |
| Gene expression levels in SubtiExpress: mfd | |
| Interactions involving this protein in SubtInteract: Mfd | |
| MW, pI | 133 kDa, 5.367 |
| Gene length, protein length | 3531 bp, 1177 aa |
| Immediate neighbours | fin, spoVT |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 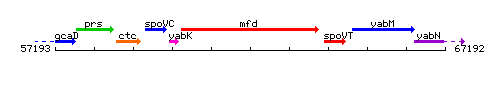
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, transcription
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00550
Phenotypes of a mutant
- in an mfd knock-out, the cell's ability to accumulate adaptive mutations in stationary phase is depressed. PubMed
- increased UV-induced mutagenesis via PolY1/ PolY2-mediated translesion synthesis PubMed
- the mutation suppresses the mucoid phenotype of motA or motB mutants PubMed
Database entries
- BsubCyc: BSU00550
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- promotes strand-specific DNA repair by displacing RNA polymerase stalled at a nucleotide lesion and directing the (A)BC excinuclease to the RNA damage site
- is required for roadblock transcription repression by transcription factors with binding sites downstream of the promoter (as for CcpA PubMed and CodY PubMed)
- required for the processing of genetic damage during sporulation PubMed
- Protein family:
- Paralogous protein(s): RecG
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU00550
- Structure:
- UniProt: P37474
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- expressed throughout growth and sporulation, during sporulation both in the mother cell and the forespore PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP1167 (del ermC), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion: GP1510 (spc, based on pGP1870, pGP1389-derivative ), available in Jörg Stülke's lab
- YFP fusion: GP1511 (spc, based on pGP1871, pGP1389-derivative ), available in Jörg Stülke's lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Fabian Commichau's lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Evan T Graves, Camille Duboc, Jun Fan, François Stransky, Mathieu Leroux-Coyau, Terence R Strick
A dynamic DNA-repair complex observed by correlative single-molecule nanomanipulation and fluorescence.
Nat Struct Mol Biol: 2015, 22(6);452-7
[PubMed:25961799]
[WorldCat.org]
[DOI]
(I p)
Samuel Million-Weaver, Ariana N Samadpour, Daniela A Moreno-Habel, Patrick Nugent, Mitchell J Brittnacher, Eli Weiss, Hillary S Hayden, Samuel I Miller, Ivan Liachko, Houra Merrikh
An underlying mechanism for the increased mutagenesis of lagging-strand genes in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2015, 112(10);E1096-105
[PubMed:25713353]
[WorldCat.org]
[DOI]
(I p)
Hirokazu Suzuki, Jyumpei Kobayashi, Keisuke Wada, Megumi Furukawa, Katsumi Doi
Thermoadaptation-directed enzyme evolution in an error-prone thermophile derived from Geobacillus kaustophilus HTA426.
Appl Environ Microbiol: 2015, 81(1);149-58
[PubMed:25326311]
[WorldCat.org]
[DOI]
(I p)
Jia Mun Chan, Sarah B Guttenplan, Daniel B Kearns
Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis.
J Bacteriol: 2014, 196(4);740-53
[PubMed:24296669]
[WorldCat.org]
[DOI]
(I p)
Fernando H Ramírez-Guadiana, Rocío Del Carmen Barajas-Ornelas, Víctor M Ayala-García, Ronald E Yasbin, Eduardo Robleto, Mario Pedraza-Reyes
Transcriptional coupling of DNA repair in sporulating Bacillus subtilis cells.
Mol Microbiol: 2013, 90(5);1088-99
[PubMed:24118570]
[WorldCat.org]
[DOI]
(I p)
Kévin Howan, Abigail J Smith, Lars F Westblade, Nicolas Joly, Wilfried Grange, Sylvain Zorman, Seth A Darst, Nigel J Savery, Terence R Strick
Initiation of transcription-coupled repair characterized at single-molecule resolution.
Nature: 2012, 490(7420);431-4
[PubMed:22960746]
[WorldCat.org]
[DOI]
(I p)
Holly Anne Martin, Mario Pedraza-Reyes, Ronald E Yasbin, Eduardo A Robleto
Transcriptional de-repression and Mfd are mutagenic in stressed Bacillus subtilis cells.
J Mol Microbiol Biotechnol: 2011, 21(1-2);45-58
[PubMed:22248542]
[WorldCat.org]
[DOI]
(I p)
Katrin Gunka, Stefan Tholen, Jan Gerwig, Christina Herzberg, Jörg Stülke, Fabian M Commichau
A high-frequency mutation in Bacillus subtilis: requirements for the decryptification of the gudB glutamate dehydrogenase gene.
J Bacteriol: 2012, 194(5);1036-44
[PubMed:22178973]
[WorldCat.org]
[DOI]
(I p)
Olivier Delumeau, François Lecointe, Jan Muntel, Alain Guillot, Eric Guédon, Véronique Monnet, Michael Hecker, Dörte Becher, Patrice Polard, Philippe Noirot
The dynamic protein partnership of RNA polymerase in Bacillus subtilis.
Proteomics: 2011, 11(15);2992-3001
[PubMed:21710567]
[WorldCat.org]
[DOI]
(I p)
Boris R Belitsky, Abraham L Sonenshein
Roadblock repression of transcription by Bacillus subtilis CodY.
J Mol Biol: 2011, 411(4);729-43
[PubMed:21699902]
[WorldCat.org]
[DOI]
(I p)
Laura Manelyte, Young-In T Kim, Abigail J Smith, Rachel M Smith, Nigel J Savery
Regulation and rate enhancement during transcription-coupled DNA repair.
Mol Cell: 2010, 40(5);714-24
[PubMed:21145481]
[WorldCat.org]
[DOI]
(I p)
Lars F Westblade, Elizabeth A Campbell, Chirangini Pukhrambam, Julio C Padovan, Bryce E Nickels, Valerie Lamour, Seth A Darst
Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction.
Nucleic Acids Res: 2010, 38(22);8357-69
[PubMed:20702425]
[WorldCat.org]
[DOI]
(I p)
Christine Pybus, Mario Pedraza-Reyes, Christian A Ross, Holly Martin, Katherine Ona, Ronald E Yasbin, Eduardo Robleto
Transcription-associated mutation in Bacillus subtilis cells under stress.
J Bacteriol: 2010, 192(13);3321-8
[PubMed:20435731]
[WorldCat.org]
[DOI]
(I p)
Christian Ross, Christine Pybus, Mario Pedraza-Reyes, Huang-Mo Sung, Ronald E Yasbin, Eduardo Robleto
Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis.
J Bacteriol: 2006, 188(21);7512-20
[PubMed:16950921]
[WorldCat.org]
[DOI]
(P p)
Alexandra M Deaconescu, Anna L Chambers, Abigail J Smith, Bryce E Nickels, Ann Hochschild, Nigel J Savery, Seth A Darst
Structural basis for bacterial transcription-coupled DNA repair.
Cell: 2006, 124(3);507-20
[PubMed:16469698]
[WorldCat.org]
[DOI]
(P p)
J M Zalieckas, L V Wray, A E Ferson, S H Fisher
Transcription-repair coupling factor is involved in carbon catabolite repression of the Bacillus subtilis hut and gnt operons.
Mol Microbiol: 1998, 27(5);1031-8
[PubMed:9535092]
[WorldCat.org]
[DOI]
(P p)
S Ayora, F Rojo, N Ogasawara, S Nakai, J C Alonso
The Mfd protein of Bacillus subtilis 168 is involved in both transcription-coupled DNA repair and DNA recombination.
J Mol Biol: 1996, 256(2);301-18
[PubMed:8594198]
[WorldCat.org]
[DOI]
(P p)
V D Filippov, E E Zagoruiko
Study of MFD in Bacillus subtilis.
Mutat Res: 1978, 52(1);49-56
[PubMed:104170]
[WorldCat.org]
[DOI]
(P p)