Difference between revisions of "TyrZ"
| Line 53: | Line 53: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * sensitive against D-Tyr | ||
| + | * a ''[[tyrS]] [[tyrZ]]'' double mutant is not viable | ||
=== Database entries === | === Database entries === | ||
| Line 68: | Line 70: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr) (according to Swiss-Prot) ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr) (according to Swiss-Prot) | + | * '''Catalyzed reaction/ biological activity:''' |
| + | ** ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr) (according to Swiss-Prot) ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr) (according to Swiss-Prot) | ||
| + | ** [[TyrZ]] is highly specific for L-Tyr, prevents misincorporation of D-Tyr into proteins, less processive than [[TyrS]] | ||
* '''Protein family:''' TyrS type 2 subfamily (according to Swiss-Prot), class-I aminoacyl-tRNA synthetase family (according to Swiss-Prot) | * '''Protein family:''' TyrS type 2 subfamily (according to Swiss-Prot), class-I aminoacyl-tRNA synthetase family (according to Swiss-Prot) | ||
| Line 113: | Line 117: | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| − | ** induced by tyrosine limitation ([[T-box]]) {{PubMed|19258532,1735721}} | + | ** induced by tyrosine limitation ([[T-box]]) {{PubMed|25733610,19258532,1735721}} |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | ** [[T-box]]: [[RNA switch]], transcriptional antitermination {{PubMed|19258532,1735721}} | + | ** [[T-box]]: [[RNA switch]], transcriptional antitermination {{PubMed|25733610,19258532,1735721}} |
** [[DtrR]]: transcriptional repression {{PubMed|25733610}} | ** [[DtrR]]: transcriptional repression {{PubMed|25733610}} | ||
| Line 123: | Line 127: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' QB53532 (aphA3), available in | + | * '''Mutant:''' QB53532 (aphA3), available in [[Jörg Stülke]]'s lab |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 141: | Line 145: | ||
=References= | =References= | ||
| − | <pubmed>19258532,1735721 , 20509597 </pubmed> | + | <pubmed>19258532,1735721 25733610, 20509597 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:08, 4 March 2015
- Description: alternative tyrosyl-tRNA synthetase, required to prevent misincorporation of D-Tyr into proteins
| Gene name | tyrZ |
| Synonyms | ipa-9r, tyrS1, tyrT |
| Essential | no |
| Product | tyrosyl-tRNA synthetase (minor) |
| Function | translation |
| Gene expression levels in SubtiExpress: tyrZ | |
| Metabolic function and regulation of this protein in SubtiPathways: tyrZ | |
| MW, pI | 46 kDa, 6.661 |
| Gene length, protein length | 1239 bp, 413 aa |
| Immediate neighbours | ywaE, ywaD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 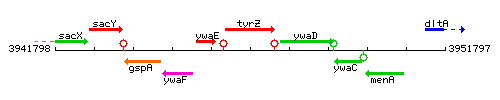
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU38460
Phenotypes of a mutant
Database entries
- BsubCyc: BSU38460
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr) (according to Swiss-Prot) ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr) (according to Swiss-Prot)
- TyrZ is highly specific for L-Tyr, prevents misincorporation of D-Tyr into proteins, less processive than TyrS
- Protein family: TyrS type 2 subfamily (according to Swiss-Prot), class-I aminoacyl-tRNA synthetase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylated on Thr-311 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU38460
- Structure:
- UniProt: P25151
- KEGG entry: [3]
- E.C. number: 6.1.1.1
Additional information
Expression and regulation
- Regulatory mechanism:
- T-box: RNA switch, transcriptional antitermination PubMed
- DtrR: transcriptional repression PubMed
- Additional information:
Biological materials
- Mutant: QB53532 (aphA3), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Rebecca N Williams-Wagner, Frank J Grundy, Medha Raina, Michael Ibba, Tina M Henkin
The Bacillus subtilis tyrZ gene encodes a highly selective tyrosyl-tRNA synthetase and is regulated by a MarR regulator and T box riboswitch.
J Bacteriol: 2015, 197(9);1624-31
[PubMed:25733610]
[WorldCat.org]
[DOI]
(I p)
Boumediene Soufi, Chanchal Kumar, Florian Gnad, Matthias Mann, Ivan Mijakovic, Boris Macek
Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis.
J Proteome Res: 2010, 9(7);3638-46
[PubMed:20509597]
[WorldCat.org]
[DOI]
(I p)
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)
T M Henkin, B L Glass, F J Grundy
Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple tRNA synthetase genes.
J Bacteriol: 1992, 174(4);1299-306
[PubMed:1735721]
[WorldCat.org]
[DOI]
(P p)