Difference between revisions of "Rny"
| Line 89: | Line 89: | ||
** [[RNase]] Y is responsible for the degradation of [[23S rRNA]], [[16S rRNA]], and mRNAs in aging spores {{PubMed|22209493}} | ** [[RNase]] Y is responsible for the degradation of [[23S rRNA]], [[16S rRNA]], and mRNAs in aging spores {{PubMed|22209493}} | ||
** [[RNase]] Y cleaves the leader of the ''[[cwlO]]'' mRNA at a stem-loop structure {{PubMed|24163346}} | ** [[RNase]] Y cleaves the leader of the ''[[cwlO]]'' mRNA at a stem-loop structure {{PubMed|24163346}} | ||
| + | ** 3' end maturation of [[rnpB|RNase P RNA]] and [[scr|scRNA]] {{PubMed|25402410}} | ||
* '''Protein family:''' Member of the HD superfamily of metal-dependent phosphohydrolases; 2',3' cyclic nucleotide phosphodiesterase family (according to Swiss-Prot) | * '''Protein family:''' Member of the HD superfamily of metal-dependent phosphohydrolases; 2',3' cyclic nucleotide phosphodiesterase family (according to Swiss-Prot) | ||
| Line 199: | Line 200: | ||
==Publications on ''B. subtilis rny''== | ==Publications on ''B. subtilis rny''== | ||
| − | <pubmed>21862575 22198292 22209493 22412379 23060960 23326572 21908660 21856853 21815947 24163346 18763711,19193632,17005971 19779461 19820159 20418391 20525796 20572937,21803996 21843271 23504012 </pubmed> | + | <pubmed>21862575 22198292 22209493 22412379 23060960 23326572 21908660 21856853 21815947 24163346 18763711,19193632,17005971 19779461 19820159 20418391 20525796 20572937,21803996 21843271 23504012 25402410</pubmed> |
==Publications on homologs from other organisms== | ==Publications on homologs from other organisms== | ||
<pubmed> 17951247 20385762 15853881 </pubmed> | <pubmed> 17951247 20385762 15853881 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:40, 18 November 2014
- Description: RNase Y, 5' end sensitive endoribonuclease, involved in the degradation/ processing of mRNA
| Gene name | rny |
| Synonyms | ymdA |
| Essential | no PubMed |
| Product | RNase Y |
| Function | RNA processing and degradation |
| Gene expression levels in SubtiExpress: rny | |
| Interactions involving this protein in SubtInteract: Rny | |
| Regulatory function of this protein in SubtiPathways: rny | |
| Metabolic function and regulation of this protein in SubtiPathways: Rny | |
| MW, pI | 58,7 kDa, 5.39 |
| Gene length, protein length | 1560 bp, 520 amino acids |
| Immediate neighbours | pbpX, ymdB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed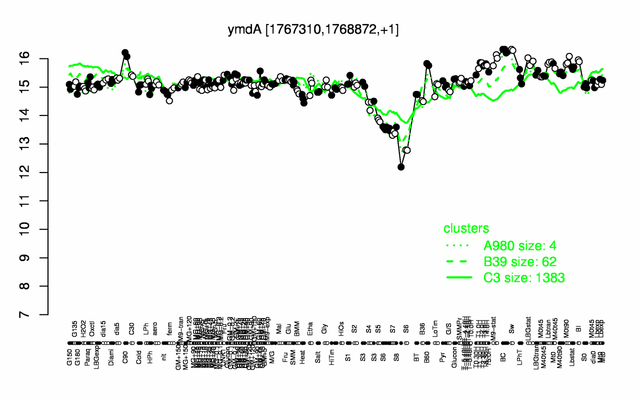
| |
Contents
Categories containing this gene/protein
Rnases, biofilm formation, membrane proteins
This gene is a member of the following regulons
Targets of RNase Y
The gene
Basic information
- Locus tag: BSU16960
Phenotypes of a mutant
- transcription profile resulting from rny depletion: GEO PubMed
- defect in spore germination PubMed
- a study from the lab of Ciaran Condon reports that rny is non-essential and that the mutant is strongly impaired in sporulation, genetic competence and many other phenotypes PubMed
Database entries
- BsubCyc: BSU16960
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- RNase Y cleaves S-box riboswitch RNAs in vivo and in vitro PubMed
- preference for 5' monophosphorylated substrate in vitro PubMed
- endonucleolytic cleavage PubMed
- required for the processing of the gapA operon mRNA PubMed
- cleavage activity appears sensitive to downstream secondary structure PubMed
- RNase Y initiates the degradation of rpsO mRNA PubMed
- RNase Y is responsible for the degradation of 23S rRNA, 16S rRNA, and mRNAs in aging spores PubMed
- RNase Y cleaves the leader of the cwlO mRNA at a stem-loop structure PubMed
- 3' end maturation of RNase P RNA and scRNA PubMed
- Protein family: Member of the HD superfamily of metal-dependent phosphohydrolases; 2',3' cyclic nucleotide phosphodiesterase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity: appears sensitive to downstream secondary structure, PubMed
Database entries
- BsubCyc: BSU16960
- Structure:
- UniProt: O31774
- KEGG entry: [3]
- E.C. number: 3.1.4.16
Additional information
required for the processing of the gapA operon mRNA
Expression and regulation
- Regulation: constitutive
- Regulatory mechanism:
- Additional information:
- there is a terminator between rny and ymdB, most transcripts terminate there PubMed
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 222 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 1386 PubMed
Biological materials
- Mutant:
- 4043 (rny under p-spac control, cat), GP193 (rny under p-xyl control, cat), both available in Jörg Stülke's lab
- SSB447 (rny under P-spac control, "erm") available in Putzer lab.
- Expression vector:
- N-terminal Strep-tag, expression in E. coli, in pGP172: pGP441, available in Jörg Stülke's lab
- N-terminal Strep-tag, for SPINE, expression in B. subtilis, in pGP380: pGP775, available in Jörg Stülke's lab
- C-terminal Strep-tag, for SPINE, expression in B. subtilis, in pGP382: pGP1852, available in Jörg Stülke's lab
- Expression of RNase Y missing the N-terminal transmembrane domain (25aa) as an intein fusion in E. coli (no tag left in the purified protein) available in the Putzer lab
- wild type rny, expression in B. subtilis, in pBQ200: pGP1201, available in Jörg Stülke's lab
- there is also a series of domain constructs present in pBQ200, all available in Jörg Stülke's lab
- chromosomal expression of Rny-Strep, spc: GP1033, available in Jörg Stülke's lab
- lacZ fusion: pGP459 (in pAC7), available in Jörg Stülke's lab
- GFP fusion:
- B. subtilis 3569 (amyE:: (p-xyl rny-gfpmut1-spc)), available in Errington lab
- pGP1368 for chromosomal expression of rny-YFP, available in Jörg Stülke's lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- FLAG-tag construct: GP1030 (spc, based on pGP1331), available in Jörg Stülke's lab
- Antibody: available in van Dijl and in Jörg Stülke's lab
Labs working on this gene/protein
- Ciaran Condon, IBPC Paris, France Homepage
- Harald Putzer, IBPC Paris, France Homepage
- Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Reviews
Publications on B. subtilis rny
Laetitia Gilet, Jeanne M DiChiara, Sabine Figaro, David H Bechhofer, Ciarán Condon
Small stable RNA maturation and turnover in Bacillus subtilis.
Mol Microbiol: 2015, 95(2);270-82
[PubMed:25402410]
[WorldCat.org]
[DOI]
(I p)
David Noone, Letal I Salzberg, Eric Botella, Katrin Bäsell, Dörte Becher, Haike Antelmann, Kevin M Devine
A highly unstable transcript makes CwlO D,L-endopeptidase expression responsive to growth conditions in Bacillus subtilis.
J Bacteriol: 2014, 196(2);237-47
[PubMed:24163346]
[WorldCat.org]
[DOI]
(I p)
Sabine Figaro, Sylvain Durand, Laetitia Gilet, Nadège Cayet, Martin Sachse, Ciarán Condon
Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence.
J Bacteriol: 2013, 195(10);2340-8
[PubMed:23504012]
[WorldCat.org]
[DOI]
(I p)
Soumaya Laalami, Philippe Bessières, Anna Rocca, Léna Zig, Pierre Nicolas, Harald Putzer
Bacillus subtilis RNase Y activity in vivo analysed by tiling microarrays.
PLoS One: 2013, 8(1);e54062
[PubMed:23326572]
[WorldCat.org]
[DOI]
(I p)
Frank Bürmann, Prachi Sawant, Marc Bramkamp
Identification of interaction partners of the dynamin-like protein DynA from Bacillus subtilis.
Commun Integr Biol: 2012, 5(4);362-9
[PubMed:23060960]
[WorldCat.org]
[DOI]
(I p)
Sylvain Durand, Laetitia Gilet, Philippe Bessières, Pierre Nicolas, Ciarán Condon
Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs.
PLoS Genet: 2012, 8(3);e1002520
[PubMed:22412379]
[WorldCat.org]
[DOI]
(I p)
Einat Segev, Yoav Smith, Sigal Ben-Yehuda
RNA dynamics in aging bacterial spores.
Cell: 2012, 148(1-2);139-49
[PubMed:22209493]
[WorldCat.org]
[DOI]
(I p)
Joseph A Newman, Lorraine Hewitt, Cecilia Rodrigues, Alexandra S Solovyova, Colin R Harwood, Richard J Lewis
Dissection of the network of interactions that links RNA processing with glycolysis in the Bacillus subtilis degradosome.
J Mol Biol: 2012, 416(1);121-36
[PubMed:22198292]
[WorldCat.org]
[DOI]
(I p)
Shiyi Yao, Jamie Richards, Joel G Belasco, David H Bechhofer
Decay of a model mRNA in Bacillus subtilis by a combination of RNase J1 5' exonuclease and RNase Y endonuclease activities.
J Bacteriol: 2011, 193(22);6384-6
[PubMed:21908660]
[WorldCat.org]
[DOI]
(I p)
Gintaras Deikus, David H Bechhofer
5' End-independent RNase J1 endonuclease cleavage of Bacillus subtilis model RNA.
J Biol Chem: 2011, 286(40);34932-40
[PubMed:21862575]
[WorldCat.org]
[DOI]
(I p)
Christine Diethmaier, Nico Pietack, Katrin Gunka, Christoph Wrede, Martin Lehnik-Habrink, Christina Herzberg, Sebastian Hübner, Jörg Stülke
A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation.
J Bacteriol: 2011, 193(21);5997-6007
[PubMed:21856853]
[WorldCat.org]
[DOI]
(I p)
Patrice Bruscella, Karen Shahbabian, Soumaya Laalami, Harald Putzer
RNase Y is responsible for uncoupling the expression of translation factor IF3 from that of the ribosomal proteins L35 and L20 in Bacillus subtilis.
Mol Microbiol: 2011, 81(6);1526-41
[PubMed:21843271]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Joseph Newman, Fabian M Rothe, Alexandra S Solovyova, Cecilia Rodrigues, Christina Herzberg, Fabian M Commichau, Richard J Lewis, Jörg Stülke
RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli.
J Bacteriol: 2011, 193(19);5431-41
[PubMed:21803996]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Henrike Pförtner, Leonie Rempeters, Nico Pietack, Christina Herzberg, Jörg Stülke
The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex.
Mol Microbiol: 2010, 77(4);958-71
[PubMed:20572937]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Shiyi Yao, David H Bechhofer
Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y.
J Bacteriol: 2010, 192(13);3279-86
[PubMed:20418391]
[WorldCat.org]
[DOI]
(I p)
Jessica C Zweers, Thomas Wiegert, Jan Maarten van Dijl
Stress-responsive systems set specific limits to the overproduction of membrane proteins in Bacillus subtilis.
Appl Environ Microbiol: 2009, 75(23);7356-64
[PubMed:19820159]
[WorldCat.org]
[DOI]
(I p)
Karen Shahbabian, Ailar Jamalli, Léna Zig, Harald Putzer
RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis.
EMBO J: 2009, 28(22);3523-33
[PubMed:19779461]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Fabian M Rothe, Christina Herzberg, Eva Wagner, Daniel Hellwig, Martin Lehnik-Habrink, Elke Hammer, Uwe Völker, Jörg Stülke
Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing.
Mol Cell Proteomics: 2009, 8(6);1350-60
[PubMed:19193632]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Alison Hunt, Joy P Rawlins, Helena B Thomaides, Jeff Errington
Functional analysis of 11 putative essential genes in Bacillus subtilis.
Microbiology (Reading): 2006, 152(Pt 10);2895-2907
[PubMed:17005971]
[WorldCat.org]
[DOI]
(P p)
Publications on homologs from other organisms