Difference between revisions of "Sco"
(→References) |
|||
| (11 intermediate revisions by 3 users not shown) | |||
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || maturation of cytochrome C oxidase caa3 | |style="background:#ABCDEF;" align="center"|'''Function''' || maturation of cytochrome C oxidase caa3 | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU21750 sco] |
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=Sco Sco] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 21 kDa, 4.81 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 21 kDa, 4.81 | ||
| Line 22: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ypmR]]'', ''[[ypmP]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ypmR]]'', ''[[ypmP]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU21750 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU21750 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU21750 DNA_with_flanks] |
|- | |- | ||
|- | |- | ||
| Line 28: | Line 30: | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ypmQ_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ypmQ_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=scuA_2291703_2292284_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:sco_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU21750]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| − | + | <br/><br/><br/><br/> | |
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
<br/><br/> | <br/><br/> | ||
| Line 51: | Line 57: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU21750&redirect=T BSU21750] | ||
* '''DBTBS entry:''' no entry | * '''DBTBS entry:''' no entry | ||
| Line 83: | Line 90: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' [[Sco]]-[[CtaC]] {{PubMed|14766920}} | + | * '''[[SubtInteract|Interactions]]:''' |
| + | ** [[Sco]]-[[CtaC]] {{PubMed|14766920}} | ||
| − | * '''Localization:''' cell membrane {{PubMed|19921776}} | + | * '''[[Localization]]:''' cell membrane {{PubMed|19921776}} |
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU21750&redirect=T BSU21750] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1XZO 1XZO] | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1XZO 1XZO] | ||
| Line 103: | Line 112: | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=scuA_2291703_2292284_-1 sco] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 109: | Line 120: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 130: | Line 141: | ||
=References= | =References= | ||
| − | + | <pubmed>14680962, 14766920, 16305244, 19027886, 10837475 19921776 20232870 21333651 21945854 22036877 25192666 21069401</pubmed> | |
| − | <pubmed>14680962, 14766920, 16305244, 19027886, 10837475 19921776 20232870 21333651 21945854</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Latest revision as of 09:29, 19 September 2014
- Description: accessory lipoprotein required for assembly of the Cu(A) center of cytochrome c oxidase caa3
| Gene name | ypmQ |
| Synonyms | |
| Essential | no |
| Product | oxidoreductase |
| Function | maturation of cytochrome C oxidase caa3 |
| Gene expression levels in SubtiExpress: sco | |
| Interactions involving this protein in SubtInteract: Sco | |
| MW, pI | 21 kDa, 4.81 |
| Gene length, protein length | 579 bp, 193 aa |
| Immediate neighbours | ypmR, ypmP |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 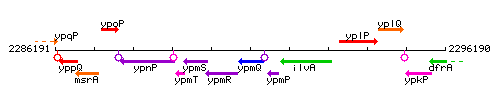
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
respiration, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU21750
Phenotypes of a mutant
Database entries
- BsubCyc: BSU21750
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): contains copper bound by two cysteines and a histidine residue PubMed
- Effectors of protein activity:
- Localization: cell membrane PubMed
Database entries
- BsubCyc: BSU21750
- Structure: 1XZO
- UniProt: P54178
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Xin Yao, Diann Andrews, Bruce C Hill
Reactivity of ligand-swapped mutants of the SCO protein from Bacillus subtilis. Isomers of the CCH metal binding motif.
Biochim Biophys Acta: 2014, 1844(12);2193-202
[PubMed:25192666]
[WorldCat.org]
[DOI]
(P p)
Mark Lai, Katherine C Yam, Diann Andrews, Bruce C Hill
Copper binding traps the folded state of the SCO protein from Bacillus subtilis.
Biochim Biophys Acta: 2012, 1824(2);292-302
[PubMed:22036877]
[WorldCat.org]
[DOI]
(P p)
Bruce C Hill, Diann Andrews
Differential affinity of BsSCO for Cu(II) and Cu(I) suggests a redox role in copper transfer to the Cu(A) center of cytochrome c oxidase.
Biochim Biophys Acta: 2012, 1817(6);948-54
[PubMed:21945854]
[WorldCat.org]
[DOI]
(P p)
Brian Bennett, Bruce C Hill
Avoiding premature oxidation during the binding of Cu(II) to a dithiolate site in BsSCO. A rapid freeze-quench EPR study.
FEBS Lett: 2011, 585(6);861-4
[PubMed:21333651]
[WorldCat.org]
[DOI]
(I p)
Gnana S Siluvai, Michiko Nakano, Mary Mayfield, Ninian J Blackburn
The essential role of the Cu(II) state of Sco in the maturation of the Cu(A) center of cytochrome oxidase: evidence from H135Met and H135SeM variants of the Bacillus subtilis Sco.
J Biol Inorg Chem: 2011, 16(2);285-97
[PubMed:21069401]
[WorldCat.org]
[DOI]
(I p)
Gnana S Siluvai, Mary Mayfield, Mark J Nilges, Serena Debeer George, Ninian J Blackburn
Anatomy of a red copper center: spectroscopic identification and reactivity of the copper centers of Bacillus subtilis Sco and its Cys-to-Ala variants.
J Am Chem Soc: 2010, 132(14);5215-26
[PubMed:20232870]
[WorldCat.org]
[DOI]
(I p)
Gnana S Siluvai, Michiko M Nakano, Mary Mayfield, Mark J Nilges, Ninian J Blackburn
H135A controls the redox activity of the Sco copper center. Kinetic and spectroscopic studies of the His135Ala variant of Bacillus subtilis Sco.
Biochemistry: 2009, 48(51);12133-44
[PubMed:19921776]
[WorldCat.org]
[DOI]
(I p)
David E Davidson, Bruce C Hill
Stability of oxidized, reduced and copper bound forms of Bacillus subtilis Sco.
Biochim Biophys Acta: 2009, 1794(2);275-81
[PubMed:19027886]
[WorldCat.org]
[DOI]
(P p)
Luisa Andruzzi, Michiko Nakano, Mark J Nilges, Ninian J Blackburn
Spectroscopic studies of metal binding and metal selectivity in Bacillus subtilis BSco, a Homologue of the Yeast Mitochondrial Protein Sco1p.
J Am Chem Soc: 2005, 127(47);16548-58
[PubMed:16305244]
[WorldCat.org]
[DOI]
(P p)
Jenny Bengtsson, Claes von Wachenfeldt, Lena Winstedt, Per Nygaard, Lars Hederstedt
CtaG is required for formation of active cytochrome c oxidase in Bacillus subtilis.
Microbiology (Reading): 2004, 150(Pt 2);415-425
[PubMed:14766920]
[WorldCat.org]
[DOI]
(P p)
Diann Andrews, Jennifer Rattenbury, Vijay Anand, Neil R Mattatall, Bruce C Hill
Expression, purification, and characterization of BsSco, an accessory protein involved in the assembly of cytochrome c oxidase in Bacillus subtilis.
Protein Expr Purif: 2004, 33(1);57-65
[PubMed:14680962]
[WorldCat.org]
[DOI]
(P p)
N R Mattatall, J Jazairi, B C Hill
Characterization of YpmQ, an accessory protein required for the expression of cytochrome c oxidase in Bacillus subtilis.
J Biol Chem: 2000, 275(37);28802-9
[PubMed:10837475]
[WorldCat.org]
[DOI]
(P p)