Difference between revisions of "OxdC"
(→References) |
|||
| Line 142: | Line 142: | ||
<pubmed>20464388 </pubmed> | <pubmed>20464388 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>12056897,18573182,17269748,14871895 ,10960116 , 19047353, 19473032 11546787 15583401 21264418 21782782 22404040 23734819 21277974 22517742 24444454 </pubmed> | + | <pubmed>12056897,18573182,17269748,14871895 ,10960116 , 19047353, 19473032 11546787 15583401 21264418 21782782 22404040 23734819 21277974 22517742 24444454 25186982 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:10, 6 September 2014
- Description: oxalate decarboxylase
| Gene name | oxdC |
| Synonyms | yvrK |
| Essential | no |
| Product | oxalate decarboxylase |
| Function | unknown |
| Gene expression levels in SubtiExpress: oxdC | |
| MW, pI | 43 kDa, 5.101 |
| Gene length, protein length | 1155 bp, 385 aa |
| Immediate neighbours | yvrJ, yvrL |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 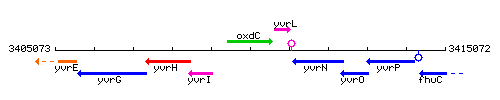
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed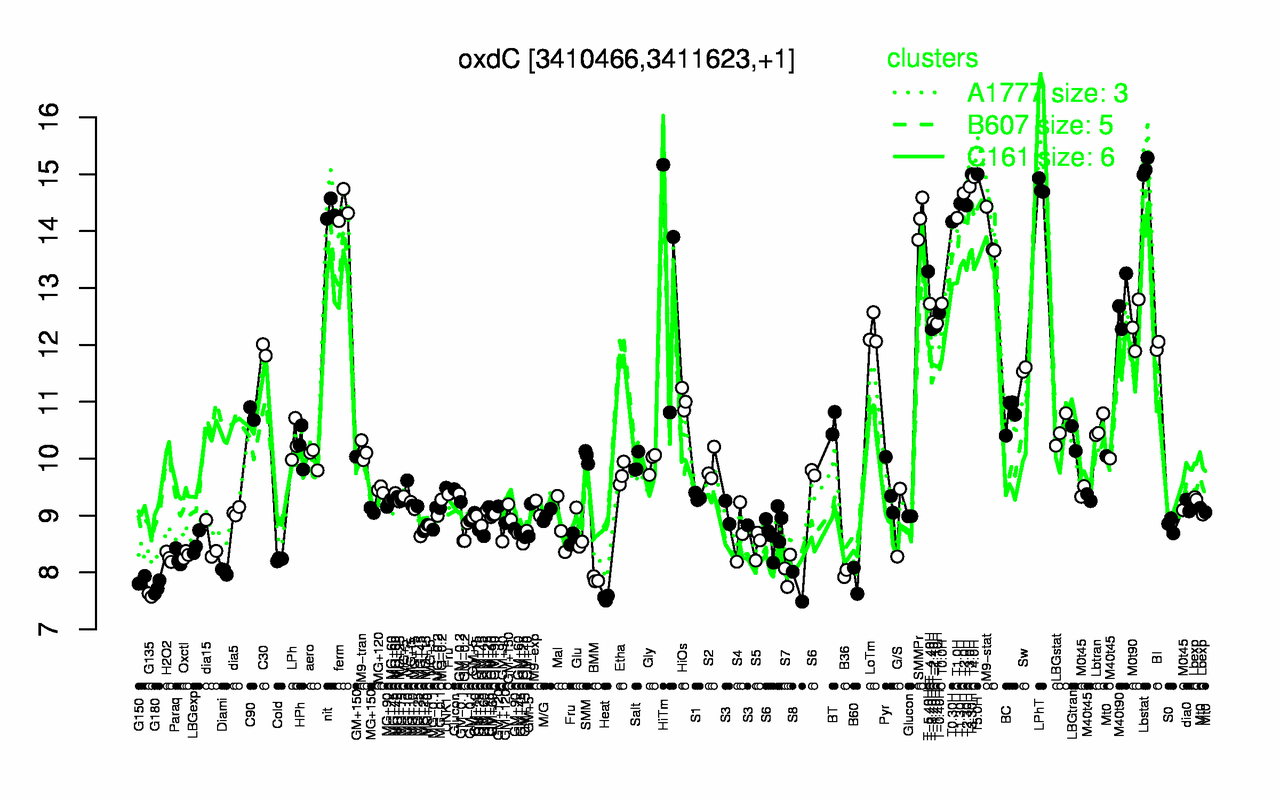
| |
Contents
Categories containing this gene/protein
acid stress proteins (controlled by YvrI-YvrHa), phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33240
Phenotypes of a mutant
Database entries
- BsubCyc: BSU33240
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Oxalate = formate + CO2 (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s): OxdD
Extended information on the protein
- Kinetic information:
- Modification:
- phosphorylated on Arg-12 PubMed
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU33240
- Structure: 1UW8
- UniProt: O34714
- KEGG entry: [2]
- E.C. number: 4.1.1.2
Additional information
Expression and regulation
- Regulation: induced by acidic growth conditions PubMed
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 193 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 4306 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 2602 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 4926 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Your additional remarks
References
Reviews
Miia R Mäkelä, Kristiina Hildén, Taina K Lundell
Oxalate decarboxylase: biotechnological update and prevalence of the enzyme in filamentous fungi.
Appl Microbiol Biotechnol: 2010, 87(3);801-14
[PubMed:20464388]
[WorldCat.org]
[DOI]
(I p)
Original publications
Eunhye Lee, Byong Chang Jeong, Yong Hyun Park, Hyeon Hoe Kim
Expression of the gene encoding oxalate decarboxylase from Bacillus subtilis and characterization of the recombinant enzyme.
BMC Res Notes: 2014, 7;598
[PubMed:25186982]
[WorldCat.org]
[DOI]
(I e)
Pablo Campomanes, Whitney F Kellett, Lindsey M Easthon, Andrew Ozarowski, Karen N Allen, Alexander Angerhofer, Ursula Rothlisberger, Nigel G J Richards
Assigning the EPR fine structure parameters of the Mn(II) centers in Bacillus subtilis oxalate decarboxylase by site-directed mutagenesis and DFT/MM calculations.
J Am Chem Soc: 2014, 136(6);2313-23
[PubMed:24444454]
[WorldCat.org]
[DOI]
(I p)
K Anbazhagan, P Sasikumar, S Gomathi, H P Priya, G S Selvam
In vitro degradation of oxalate by recombinant Lactobacillus plantarum expressing heterologous oxalate decarboxylase.
J Appl Microbiol: 2013, 115(3);880-7
[PubMed:23734819]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Benjamin T Saylor, Laurie A Reinhardt, Zhibing Lu, Mithila S Shukla, Linda Nguyen, W Wallace Cleland, Alexander Angerhofer, Karen N Allen, Nigel G J Richards
A structural element that facilitates proton-coupled electron transfer in oxalate decarboxylase.
Biochemistry: 2012, 51(13);2911-20
[PubMed:22404040]
[WorldCat.org]
[DOI]
(I p)
Mario E G Moral, Chingkuang Tu, Nigel G J Richards, David N Silverman
Membrane inlet for mass spectrometric measurement of catalysis by enzymatic decarboxylases.
Anal Biochem: 2011, 418(1);73-7
[PubMed:21782782]
[WorldCat.org]
[DOI]
(I p)
Witcha Imaram, Benjamin T Saylor, Christopher P Centonze, Nigel G J Richards, Alexander Angerhofer
EPR spin trapping of an oxalate-derived free radical in the oxalate decarboxylase reaction.
Free Radic Biol Med: 2011, 50(8);1009-15
[PubMed:21277974]
[WorldCat.org]
[DOI]
(I p)
Mario E G Moral, Chingkuang Tu, Witcha Imaram, Alexander Angerhofer, David N Silverman, Nigel G J Richards
Nitric oxide reversibly inhibits Bacillus subtilis oxalate decarboxylase.
Chem Commun (Camb): 2011, 47(11);3111-3
[PubMed:21264418]
[WorldCat.org]
[DOI]
(I p)
Ellen W Moomaw, Alexander Angerhofer, Patricia Moussatche, Andrew Ozarowski, Inés García-Rubio, Nigel G J Richards
Metal dependence of oxalate decarboxylase activity.
Biochemistry: 2009, 48(26);6116-25
[PubMed:19473032]
[WorldCat.org]
[DOI]
(I p)
Shawn R MacLellan, John D Helmann, Haike Antelmann
The YvrI alternative sigma factor is essential for acid stress induction of oxalate decarboxylase in Bacillus subtilis.
J Bacteriol: 2009, 191(3);931-9
[PubMed:19047353]
[WorldCat.org]
[DOI]
(I p)
Shawn R MacLellan, Tina Wecke, John D Helmann
A previously unidentified sigma factor and two accessory proteins regulate oxalate decarboxylase expression in Bacillus subtilis.
Mol Microbiol: 2008, 69(4);954-67
[PubMed:18573182]
[WorldCat.org]
[DOI]
(I p)
Haike Antelmann, Stefanie Töwe, Dirk Albrecht, Michael Hecker
The phosphorus source phytate changes the composition of the cell wall proteome in Bacillus subtilis.
J Proteome Res: 2007, 6(2);897-903
[PubMed:17269748]
[WorldCat.org]
[DOI]
(P p)
Clare E M Stevenson, Adam Tanner, Laura Bowater, Stephen Bornemann, David M Lawson
SAD at home: solving the structure of oxalate decarboxylase with the anomalous signal from manganese using X-ray data collected on a home source.
Acta Crystallogr D Biol Crystallogr: 2004, 60(Pt 12 Pt 2);2403-6
[PubMed:15583401]
[WorldCat.org]
[DOI]
(P p)
Victoria J Just, Clare E M Stevenson, Laura Bowater, Adam Tanner, David M Lawson, Stephen Bornemann
A closed conformation of Bacillus subtilis oxalate decarboxylase OxdC provides evidence for the true identity of the active site.
J Biol Chem: 2004, 279(19);19867-74
[PubMed:14871895]
[WorldCat.org]
[DOI]
(P p)
Ruchi Anand, Pieter C Dorrestein, Cynthia Kinsland, Tadhg P Begley, Steven E Ealick
Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
Biochemistry: 2002, 41(24);7659-69
[PubMed:12056897]
[WorldCat.org]
[DOI]
(P p)
A Tanner, L Bowater, S A Fairhurst, S Bornemann
Oxalate decarboxylase requires manganese and dioxygen for activity. Overexpression and characterization of Bacillus subtilis YvrK and YoaN.
J Biol Chem: 2001, 276(47);43627-34
[PubMed:11546787]
[WorldCat.org]
[DOI]
(P p)
A Tanner, S Bornemann
Bacillus subtilis YvrK is an acid-induced oxalate decarboxylase.
J Bacteriol: 2000, 182(18);5271-3
[PubMed:10960116]
[WorldCat.org]
[DOI]
(P p)