Difference between revisions of "GlnR"
| Line 1: | Line 1: | ||
| − | * '''Description:''' transcriptional repressor of the'' [[glnR]]-[[glnA]]'' operon <br/><br/> | + | * '''Description:''' transcriptional repressor ([[MerR family]]) of the'' [[glnR]]-[[glnA]]'' operon <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || transcription repressor | + | |style="background:#ABCDEF;" align="center"| '''Product''' || transcription repressor ([[MerR family]]) |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of glutamine synthesis | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of glutamine synthesis | ||
| Line 66: | Line 66: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 76: | Line 73: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' [[MerR family]] |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
Revision as of 13:17, 22 June 2014
- Description: transcriptional repressor (MerR family) of the glnR-glnA operon
| Gene name | glnR |
| Synonyms | |
| Essential | no |
| Product | transcription repressor (MerR family) |
| Function | regulation of glutamine synthesis |
| Gene expression levels in SubtiExpress: glnR | |
| Interactions involving this protein in SubtInteract: GlnR | |
| Metabolic function and regulation of this protein in SubtiPathways: glnR | |
| MW, pI | 15 kDa, 9.731 |
| Gene length, protein length | 405 bp, 135 aa |
| Immediate neighbours | ynbB, glnA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 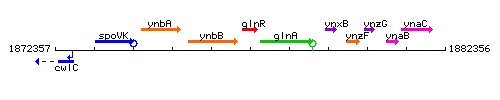
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, glutamate metabolism, transcription factors and their control
This gene is a member of the following regulons
The GlnR regulon:
The gene
Basic information
- Locus tag: BSU17450
Phenotypes of a mutant
Database entries
- BsubCyc: BSU17450
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: MerR family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Effectors of protein activity: activity is enhanced upon interaction of the C-terminal domain with GlnA PubMed
Database entries
- BsubCyc: BSU17450
- Structure:
- UniProt: P37582
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Susan Fisher, Boston, USA homepage
Your additional remarks
References
Reviews
Original publications
Ksenia Fedorova, Airat Kayumov, Kathrin Woyda, Olga Ilinskaja, Karl Forchhammer
Transcription factor TnrA inhibits the biosynthetic activity of glutamine synthetase in Bacillus subtilis.
FEBS Lett: 2013, 587(9);1293-8
[PubMed:23535029]
[WorldCat.org]
[DOI]
(I p)
Susan H Fisher, Lewis V Wray
Novel trans-Acting Bacillus subtilis glnA mutations that derepress glnRA expression.
J Bacteriol: 2009, 191(8);2485-92
[PubMed:19233925]
[WorldCat.org]
[DOI]
(I p)
Lewis V Wray, Susan H Fisher
Bacillus subtilis GlnR contains an autoinhibitory C-terminal domain required for the interaction with glutamine synthetase.
Mol Microbiol: 2008, 68(2);277-85
[PubMed:18331450]
[WorldCat.org]
[DOI]
(I p)
Susan H Fisher, Lewis V Wray
Bacillus subtilis glutamine synthetase regulates its own synthesis by acting as a chaperone to stabilize GlnR-DNA complexes.
Proc Natl Acad Sci U S A: 2008, 105(3);1014-9
[PubMed:18195355]
[WorldCat.org]
[DOI]
(I p)
Jill M Zalieckas, Lewis V Wray, Susan H Fisher
Cross-regulation of the Bacillus subtilis glnRA and tnrA genes provides evidence for DNA binding site discrimination by GlnR and TnrA.
J Bacteriol: 2006, 188(7);2578-85
[PubMed:16547045]
[WorldCat.org]
[DOI]
(P p)
Jaclyn L Brandenburg, Lewis V Wray, Lars Beier, Hanne Jarmer, Hans H Saxild, Susan H Fisher
Roles of PucR, GlnR, and TnrA in regulating expression of the Bacillus subtilis ure P3 promoter.
J Bacteriol: 2002, 184(21);6060-4
[PubMed:12374841]
[WorldCat.org]
[DOI]
(P p)
L V Wray, A E Ferson, S H Fisher
Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H.
J Bacteriol: 1997, 179(17);5494-501
[PubMed:9287005]
[WorldCat.org]
[DOI]
(P p)
L V Wray, A E Ferson, K Rohrer, S H Fisher
TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1996, 93(17);8841-5
[PubMed:8799114]
[WorldCat.org]
[DOI]
(P p)
S W Brown, A L Sonenshein
Autogenous regulation of the Bacillus subtilis glnRA operon.
J Bacteriol: 1996, 178(8);2450-4
[PubMed:8636055]
[WorldCat.org]
[DOI]
(P p)
H J Schreier, S W Brown, K D Hirschi, J F Nomellini, A L Sonenshein
Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene.
J Mol Biol: 1989, 210(1);51-63
[PubMed:2573733]
[WorldCat.org]
[DOI]
(P p)
M A Strauch, A I Aronson, S W Brown, H J Schreier, A L Sonenhein
Sequence of the Bacillus subtilis glutamine synthetase gene region.
Gene: 1988, 71(2);257-65
[PubMed:2906311]
[WorldCat.org]
[DOI]
(P p)