Difference between revisions of "CodY"
| Line 132: | Line 132: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
** the intracellular concentration of CodY is about 2.5 myM (according to {{PubMed|20408793}}) | ** the intracellular concentration of CodY is about 2.5 myM (according to {{PubMed|20408793}}) | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 955 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 3409 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 09:46, 17 April 2014
- Description: regulation of a large regulon (more than 100 genes and operons) in response to branched-chain amino acid limitation
| Gene name | codY |
| Synonyms | |
| Essential | no |
| Product | transcriptional pleiotropic repressor |
| Function | regulation of a large regulon in response to
branched-chain amino acid limitation to the presence of branched-chain amino acids |
| Gene expression levels in SubtiExpress: codY | |
| Interactions involving this protein in SubtInteract: CodY | |
| Metabolic function and regulation of this protein in SubtiPathways: codY | |
| MW, pI | 28 kDa, 4.75 |
| Gene length, protein length | 777 bp, 259 aa |
| Immediate neighbours | clpY, flgB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 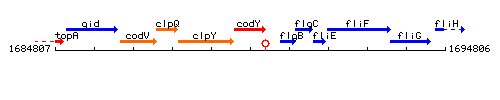
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
transcription factors and their control, regulators of core metabolism, phosphoproteins
This gene is a member of the following regulons
The CodY regulon
The gene
Basic information
- Locus tag: BSU16170
Phenotypes of a mutant
- no swarming motility on B medium. PubMed
- the mutation suppresses the mucoid phenotype of motA or motB mutants due to loss of DegU phosphorylation and concomitant reduced expression of the capB-capC-capA-capE operon PubMed
- inactivation of codY suppresses the requirement of a relA sasA sasB triple mutant for branched chain amino acids, methionine and threonine PubMed
Database entries
- BsubCyc: BSU16170
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: codY family (according to Swiss-Prot)
- Paralogous protein(s):
Genes/ operons controlled by CodY
Extended information on the protein
- Kinetic information:
- Domains: contains a GAF domain (ligand binding domain)
- Modification: phosphorylation on Ser-215 PubMed
- Effectors of protein activity: GTP and branched chained amino acids (BCAA) increase the affinity of CodY for its DNA target sequences PubMed
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU16170
- UniProt: P39779
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- GP566, available in Stülke lab
- a codY::erm mutant is available in Linc Sonenshein's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Tony Wilkinson, York University, U.K. homepage
Oscar Kuipers, University of Groningen, The Netherlands Homepage
Your additional remarks
References
Reviews
The CodY regulon
Boris R Belitsky, Abraham L Sonenshein
Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution.
Proc Natl Acad Sci U S A: 2013, 110(17);7026-31
[PubMed:23569278]
[WorldCat.org]
[DOI]
(I p)
Boris R Belitsky, Abraham L Sonenshein
Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis.
J Bacteriol: 2008, 190(4);1224-36
[PubMed:18083814]
[WorldCat.org]
[DOI]
(I p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
Original Publications