Difference between revisions of "Pgi"
| Line 120: | Line 120: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 5183 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 17347 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 09:45, 17 April 2014
- Description: glucose 6-phosphate isomerase, glycolytic / gluconeogenic enzyme
| Gene name | pgi |
| Synonyms | yugL |
| Essential | no |
| Product | glucose-6-phosphate isomerase |
| Function | enzyme in glycolysis / gluconeogenesis |
| Gene expression levels in SubtiExpress: pgi | |
| Metabolic function and regulation of this protein in SubtiPathways: pgi | |
| MW, pI | 50.4 kDa, 4.85 |
| Gene length, protein length | 1353 bp, 451 amino acids |
| Immediate neighbours | yugM, yugK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 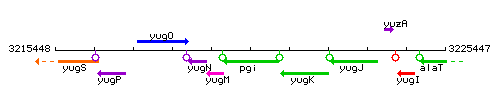
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU31350
Phenotypes of a mutant
Database entries
- BsubCyc: BSU31350
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: D-glucose 6-phosphate = D-fructose 6-phosphate (according to Swiss-Prot) D-glucose 6-phosphate = D-fructose 6-phosphate
- Protein family: GPI family (according to Swiss-Prot) GPI family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification: phosphorylation on Thr-39 PubMed
- Effectors of protein activity: competitively inhibited by 6-phosphogluconate (in B.caldotenax, B.stearothermophilus) PubMed
- Localization:
- cytoplasm (according to Swiss-Prot), cytoplasm
Database entries
- BsubCyc: BSU31350
- UniProt: P80860
- KEGG entry: [3]
- E.C. number: 5.3.1.9
Additional information
- extensive information on the structure and enzymatic properties of Pgi can be found at Proteopedia
Expression and regulation
- Regulation: constitutively expressed PubMed
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 5183 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 17347 PubMed
Biological materials
- Mutant: GP508 (spc), available in Jörg Stülke's lab, PubMed
- Expression vector:
- pGP398 (N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- lacZ fusion: pGP510 (in pAC6), available in Jörg Stülke's lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany homepage
Your additional remarks
References