Difference between revisions of "MurC"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 58: | Line 54: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | + | * essential [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | |
| − | essential [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | + | * murC is dispensable in wall-less L-forms {{PubMed|24704074}} |
=== Database entries === | === Database entries === | ||
| Line 69: | Line 65: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 87: | Line 80: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 119: | Line 112: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=murC_3048177_3049475_-1 murC] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=murC_3048177_3049475_-1 murC] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 147: | Line 140: | ||
=References= | =References= | ||
| − | <pubmed>18063720,8733232 , 16479537 </pubmed> | + | <pubmed>18063720,8733232 , 24704074 16479537 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:39, 8 April 2014
- Description: UDP-N-acetylmuramoyl-L-alanine synthetase
| Gene name | murC |
| Synonyms | ytxF |
| Essential | yes PubMed |
| Product | UDP-N-acetylmuramoyl-L-alanine synthetase |
| Function | peptidoglycan precursor biosynthesis |
| Gene expression levels in SubtiExpress: murC | |
| Metabolic function and regulation of this protein in SubtiPathways: murC | |
| MW, pI | 48 kDa, 5.307 |
| Gene length, protein length | 1296 bp, 432 aa |
| Immediate neighbours | ytxG, sftA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 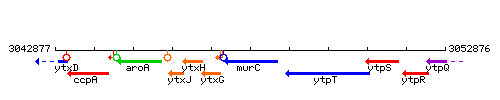
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed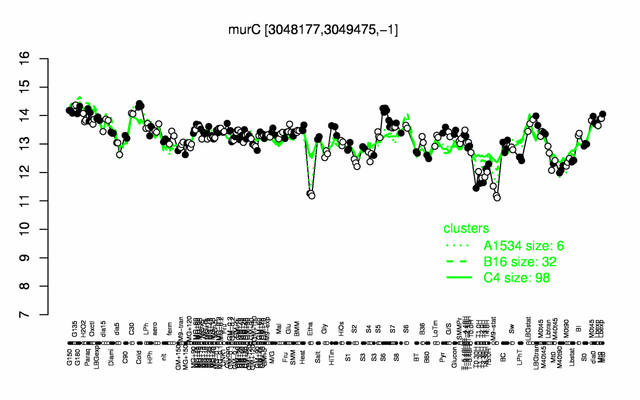
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29790
Phenotypes of a mutant
Database entries
- BsubCyc: BSU29790
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + UDP-N-acetylmuramate + L-alanine = ADP + phosphate + UDP-N-acetylmuramoyl-L-alanine (according to Swiss-Prot)
- Protein family: murCDEF family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- cytoplasm (Homogeneous) PubMed
Database entries
- BsubCyc: BSU29790
- Structure:
- UniProt: P40778
- KEGG entry: [3]
- E.C. number: 6.3.2.8
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Yoshikazu Kawai, Romain Mercier, Jeff Errington
Bacterial cell morphogenesis does not require a preexisting template structure.
Curr Biol: 2014, 24(8);863-7
[PubMed:24704074]
[WorldCat.org]
[DOI]
(I p)
Jean van Heijenoort
Lipid intermediates in the biosynthesis of bacterial peptidoglycan.
Microbiol Mol Biol Rev: 2007, 71(4);620-35
[PubMed:18063720]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
D Varón, M S Brody, C W Price
Bacillus subtilis operon under the dual control of the general stress transcription factor sigma B and the sporulation transcription factor sigma H.
Mol Microbiol: 1996, 20(2);339-50
[PubMed:8733232]
[WorldCat.org]
[DOI]
(P p)