Difference between revisions of "OdhA"
| Line 59: | Line 59: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU19370&redirect=T BSU19370] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/odhAB.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/odhAB.html] | ||
| Line 95: | Line 96: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU19370&redirect=T BSU19370] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2jgd 2JGD] (from ''Escherichia coli'', 38% identity, 55% similarity) {{PubMed|17367808}} | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2jgd 2JGD] (from ''Escherichia coli'', 38% identity, 55% similarity) {{PubMed|17367808}} | ||
Revision as of 13:55, 2 April 2014
- Description: 2-oxoglutarate dehydrogenase (E1 subunit)
| Gene name | odhA |
| Synonyms | citK |
| Essential | no |
| Product | 2-oxoglutarate dehydrogenase (E1 subunit) |
| Function | TCA cycle |
| Gene expression levels in SubtiExpress: odhA | |
| Interactions involving this protein in SubtInteract: OdhA | |
| Metabolic function and regulation of this protein in SubtiPathways: odhA | |
| MW, pI | 105 kDa, 5.871 |
| Gene length, protein length | 2823 bp, 941 aa |
| Immediate neighbours | odhB, yojO |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 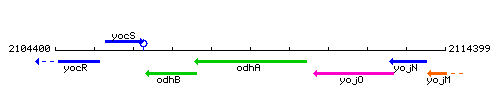
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed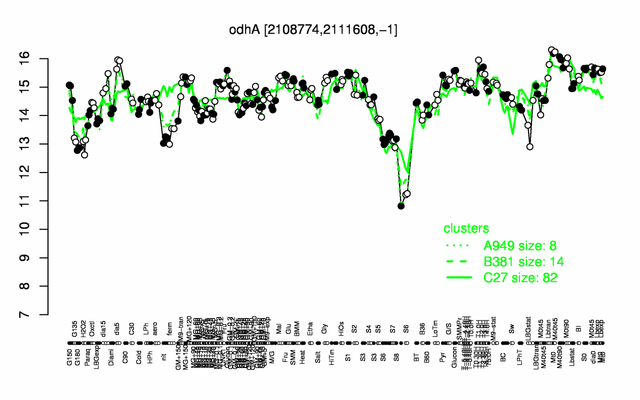
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU19370
Phenotypes of a mutant
Database entries
- BsubCyc: BSU19370
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-oxoglutarate + [dihydrolipoyllysine-residue succinyltransferase] lipoyllysine = [dihydrolipoyllysine-residue succinyltransferase] S-succinyldihydrolipoyllysine + CO2 (according to Swiss-Prot)
- Protein family: 2-oxoacid dehydrogenase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- phosphorylated on Arg-66 and Arg-621 PubMed
- Effectors of protein activity:
Database entries
- BsubCyc: BSU19370
- UniProt: P23129
- KEGG entry: [3]
- E.C. number: 1.2.4.2
Additional information
- extensive information on the structure and enzymatic properties of 2-oxoglutarate dehydrogenase can be found at Proteopedia
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- GP671 (spc), GP684 (cat), GP1274 (cat) available in Jörg Stülke's lab
- GP1276 (odhA-odhB::cat), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
K F Sheu, J P Blass
The alpha-ketoglutarate dehydrogenase complex.
Ann N Y Acad Sci: 1999, 893;61-78
[PubMed:10672230]
[WorldCat.org]
[DOI]
(P p)
Original publications
Andreas Schmidt, Débora Broch Trentini, Silvia Spiess, Jakob Fuhrmann, Gustav Ammerer, Karl Mechtler, Tim Clausen
Quantitative phosphoproteomics reveals the role of protein arginine phosphorylation in the bacterial stress response.
Mol Cell Proteomics: 2014, 13(2);537-50
[PubMed:24263382]
[WorldCat.org]
[DOI]
(I p)
Bogumiła C Marciniak, Monika Pabijaniak, Anne de Jong, Robert Dűhring, Gerald Seidel, Wolfgang Hillen, Oscar P Kuipers
High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis.
BMC Genomics: 2012, 13;401
[PubMed:22900538]
[WorldCat.org]
[DOI]
(I e)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Frederik M Meyer, Jan Gerwig, Elke Hammer, Christina Herzberg, Fabian M Commichau, Uwe Völker, Jörg Stülke
Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon.
Metab Eng: 2011, 13(1);18-27
[PubMed:20933603]
[WorldCat.org]
[DOI]
(I p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Ciarán Condon, Jordi Rourera, Dominique Brechemier-Baey, Harald Putzer
Ribonuclease M5 has few, if any, mRNA substrates in Bacillus subtilis.
J Bacteriol: 2002, 184(10);2845-9
[PubMed:11976317]
[WorldCat.org]
[DOI]
(P p)
O Resnekov, L Melin, P Carlsson, M Mannerlöv, A von Gabain, L Hederstedt
Organization and regulation of the Bacillus subtilis odhAB operon, which encodes two of the subenzymes of the 2-oxoglutarate dehydrogenase complex.
Mol Gen Genet: 1992, 234(2);285-96
[PubMed:1508153]
[WorldCat.org]
[DOI]
(P p)
P Carlsson, L Hederstedt
Genetic characterization of Bacillus subtilis odhA and odhB, encoding 2-oxoglutarate dehydrogenase and dihydrolipoamide transsuccinylase, respectively.
J Bacteriol: 1989, 171(7);3667-72
[PubMed:2500417]
[WorldCat.org]
[DOI]
(P p)