Difference between revisions of "QueC"
(→References) |
|||
| Line 134: | Line 134: | ||
=References= | =References= | ||
| − | <pubmed>18491386,14660578,17384645 19285444 , 19354300 24003028 21410253,21375305</pubmed> | + | <pubmed>18491386,14660578,17384645 19285444 , 19354300 24003028 21410253,21375305 24663240 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:24, 26 March 2014
- Description: synthesis of the modified ribonucleotide queuosine
| Gene name | queC |
| Synonyms | ykvJ |
| Essential | no |
| Product | unknown |
| Function | tRNA modification |
| Gene expression levels in SubtiExpress: queC | |
| MW, pI | 24 kDa, 5.147 |
| Gene length, protein length | 657 bp, 219 aa |
| Immediate neighbours | ykvI, queD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed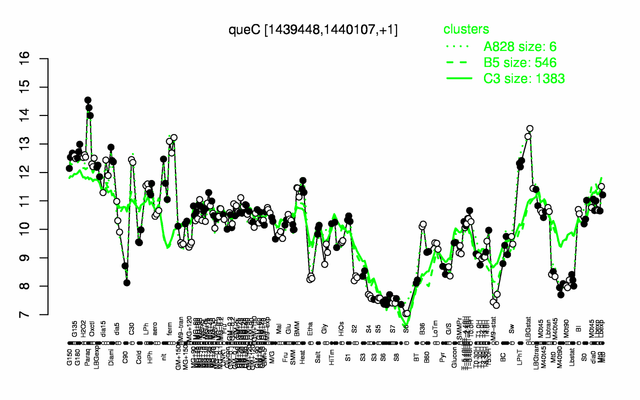
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU13720
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: queC family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 3BL5
- UniProt: O31675
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- repressed in the presence of queuosine (preQ1 riboswitch) PubMed
- Regulatory mechanism:
- preQ1 riboswitch: transcriptional antitermination in the absence of queuosine PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Zhou Gong, Yunjie Zhao, Changjun Chen, Yong Duan, Yi Xiao
Insights into ligand binding to PreQ1 Riboswitch Aptamer from molecular dynamics simulations.
PLoS One: 2014, 9(3);e92247
[PubMed:24663240]
[WorldCat.org]
[DOI]
(I e)
Krishna C Suddala, Arlie J Rinaldi, Jun Feng, Anthony M Mustoe, Catherine D Eichhorn, Joseph A Liberman, Joseph E Wedekind, Hashim M Al-Hashimi, Charles L Brooks, Nils G Walter
Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure.
Nucleic Acids Res: 2013, 41(22);10462-75
[PubMed:24003028]
[WorldCat.org]
[DOI]
(I p)
Qi Zhang, Mijeong Kang, Robert D Peterson, Juli Feigon
Comparison of solution and crystal structures of preQ1 riboswitch reveals calcium-induced changes in conformation and dynamics.
J Am Chem Soc: 2011, 133(14);5190-3
[PubMed:21410253]
[WorldCat.org]
[DOI]
(I p)
Jun Feng, Nils G Walter, Charles L Brooks
Cooperative and directional folding of the preQ1 riboswitch aptamer domain.
J Am Chem Soc: 2011, 133(12);4196-9
[PubMed:21375305]
[WorldCat.org]
[DOI]
(I p)
Reid M McCarty, Arpád Somogyi, Guangxin Lin, Neil E Jacobsen, Vahe Bandarian
The deazapurine biosynthetic pathway revealed: in vitro enzymatic synthesis of PreQ(0) from guanosine 5'-triphosphate in four steps.
Biochemistry: 2009, 48(18);3847-52
[PubMed:19354300]
[WorldCat.org]
[DOI]
(I p)
Mijeong Kang, Robert Peterson, Juli Feigon
Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA.
Mol Cell: 2009, 33(6);784-90
[PubMed:19285444]
[WorldCat.org]
[DOI]
(I p)
Nenad Cicmil, Raven H Huang
Crystal structure of QueC from Bacillus subtilis: an enzyme involved in preQ1 biosynthesis.
Proteins: 2008, 72(3);1084-8
[PubMed:18491386]
[WorldCat.org]
[DOI]
(I p)
Adam Roth, Wade C Winkler, Elizabeth E Regulski, Bobby W K Lee, Jinsoo Lim, Inbal Jona, Jeffrey E Barrick, Ankita Ritwik, Jane N Kim, Rüdiger Welz, Dirk Iwata-Reuyl, Ronald R Breaker
A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain.
Nat Struct Mol Biol: 2007, 14(4);308-17
[PubMed:17384645]
[WorldCat.org]
[DOI]
(P p)
John S Reader, David Metzgar, Paul Schimmel, Valérie de Crécy-Lagard
Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine.
J Biol Chem: 2004, 279(8);6280-5
[PubMed:14660578]
[WorldCat.org]
[DOI]
(P p)