Difference between revisions of "RsbP"
| Line 18: | Line 18: | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=RsbP RsbP] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=RsbP RsbP] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/ | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=rsbP rsbP]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 45 kDa, 4.827 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 45 kDa, 4.827 | ||
Revision as of 11:42, 7 January 2014
- Description: protein serine phosphatase, energy PP2C, dephosphorylates RsbV
| Gene name | rsbP |
| Synonyms | yvfP |
| Essential | no |
| Product | protein serine phosphatase, energy PP2C |
| Function | control of SigB activity |
| Gene expression levels in SubtiExpress: rsbP | |
| Interactions involving this protein in SubtInteract: RsbP | |
| Metabolic function and regulation of this protein in SubtiPathways: rsbP | |
| MW, pI | 45 kDa, 4.827 |
| Gene length, protein length | 1209 bp, 403 aa |
| Immediate neighbours | rsbQ, ganB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 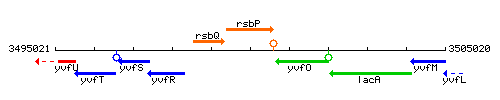
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed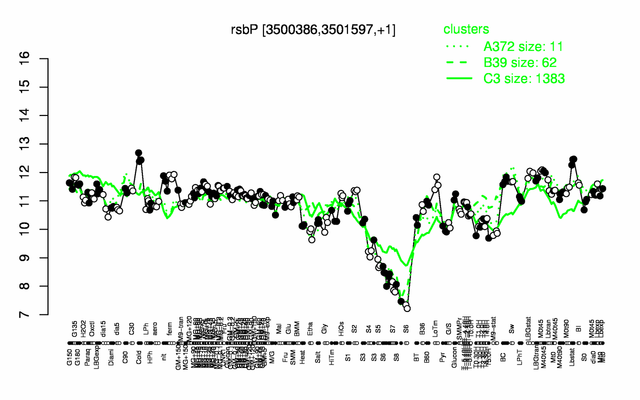
| |
Contents
Categories containing this gene/protein
protein modification, sigma factors and their control
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU34110
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: dephosphorylation of RsbV in response to red light, this results in SigB activation PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains: N-terminal PAS domain, central coiled-coil domain, C-terminal PP2C phosphatase domain
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- activity is stimulated upon interaction of the RsbP oligomer with RsbQ PubMed
Database entries
- Structure:
- UniProt: O07014
- KEGG entry: [2]
- E.C. number: 3.1.3.3
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Chet Price, Davis, USA homepage
Your additional remarks
References
Reviews
Jon Marles-Wright, Richard J Lewis
The stressosome: molecular architecture of a signalling hub.
Biochem Soc Trans: 2010, 38(4);928-33
[PubMed:20658979]
[WorldCat.org]
[DOI]
(I p)
Original publications
Additional publications: PubMed
Locke JC, Young JW, Fontes M, Hernández Jiménez MJ, Elowitz MB Stochastic pulse regulation in bacterial stress response. Science. 2011 334:366-369. PubMed:21979936
Marcela Avila-Pérez, Jeroen B van der Steen, Remco Kort, Klaas J Hellingwerf
Red light activates the sigmaB-mediated general stress response of Bacillus subtilis via the energy branch of the upstream signaling cascade.
J Bacteriol: 2010, 192(3);755-62
[PubMed:19948797]
[WorldCat.org]
[DOI]
(I p)
Masatomo Makino, Shinpei Kondo, Tomonori Kaneko, Seiki Baba, Kunio Hirata, Takashi Kumasaka
Expression, crystallization and preliminary crystallographic analysis of the PAS domain of RsbP, a stress-response phosphatase from Bacillus subtilis.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2009, 65(Pt 6);559-61
[PubMed:19478430]
[WorldCat.org]
[DOI]
(I p)
Margaret S Brody, Valley Stewart, Chester W Price
Bypass suppression analysis maps the signalling pathway within a multidomain protein: the RsbP energy stress phosphatase 2C from Bacillus subtilis.
Mol Microbiol: 2009, 72(5);1221-34
[PubMed:19432806]
[WorldCat.org]
[DOI]
(I p)
Shuyu Zhang, W G Haldenwang
Contributions of ATP, GTP, and redox state to nutritional stress activation of the Bacillus subtilis sigmaB transcription factor.
J Bacteriol: 2005, 187(22);7554-60
[PubMed:16267279]
[WorldCat.org]
[DOI]
(P p)
Tomonori Kaneko, Nobuo Tanaka, Takashi Kumasaka
Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis.
Protein Sci: 2005, 14(2);558-65
[PubMed:15632289]
[WorldCat.org]
[DOI]
(P p)
Gudrun Holtmann, Matthias Brigulla, Leif Steil, Alexandra Schütz, Karsta Barnekow, Uwe Völker, Erhard Bremer
RsbV-independent induction of the SigB-dependent general stress regulon of Bacillus subtilis during growth at high temperature.
J Bacteriol: 2004, 186(18);6150-8
[PubMed:15342585]
[WorldCat.org]
[DOI]
(P p)
Matthias Brigulla, Tamara Hoffmann, Andrea Krisp, Andrea Völker, Erhard Bremer, Uwe Völker
Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation.
J Bacteriol: 2003, 185(15);4305-14
[PubMed:12867438]
[WorldCat.org]
[DOI]
(P p)
M S Brody, K Vijay, C W Price
Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the sigma(B) transcription factor in Bacillus subtilis.
J Bacteriol: 2001, 183(21);6422-8
[PubMed:11591687]
[WorldCat.org]
[DOI]
(P p)
K Vijay, M S Brody, E Fredlund, C W Price
A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis.
Mol Microbiol: 2000, 35(1);180-8
[PubMed:10632888]
[WorldCat.org]
[DOI]
(P p)