Difference between revisions of "XlyA"
(→Extended information on the protein) |
(→Basic information/ Evolution) |
||
| Line 65: | Line 65: | ||
* '''Catalyzed reaction/ biological activity:''' Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides (according to Swiss-Prot) | * '''Catalyzed reaction/ biological activity:''' Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides (according to Swiss-Prot) | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' |
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' [[CwlA]], [[CwlH]], [[XlyB]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
Revision as of 14:57, 27 December 2013
- Description: N-acetylmuramoyl-L-alanine amidase
| Gene name | xlyA |
| Synonyms | |
| Essential | no |
| Product | N-acetylmuramoyl-L-alanine amidase |
| Function | PBSX prophage-mediated lysis |
| Gene expression levels in SubtiExpress: xlyA | |
| MW, pI | 31 kDa, 5.342 |
| Gene length, protein length | 891 bp, 297 aa |
| Immediate neighbours | xhlB, spoIISB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 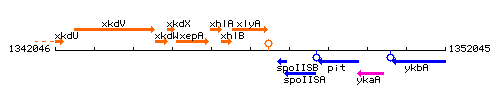
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed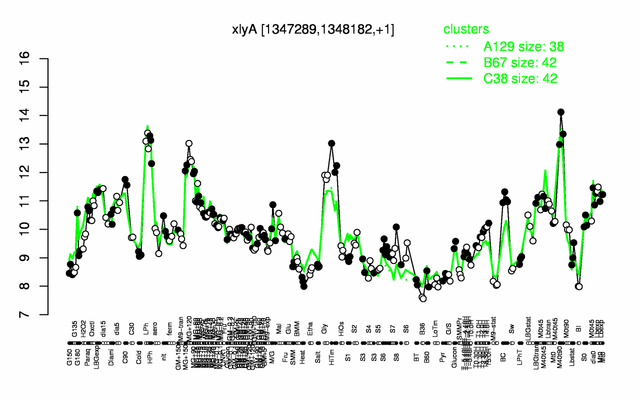
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover, PBSX prophage
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU12810
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides (according to Swiss-Prot)
- Protein family:
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- extracellular (no signal peptide) PubMed
Database entries
- UniProt: P39800
- KEGG entry: [2]
- E.C. number: 3.5.1.28
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Lieh Yoon Low, Chen Yang, Marta Perego, Andrei Osterman, Robert Liddington
Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins.
J Biol Chem: 2011, 286(39);34391-403
[PubMed:21816821]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
S Krogh, S T Jørgensen, K M Devine
Lysis genes of the Bacillus subtilis defective prophage PBSX.
J Bacteriol: 1998, 180(8);2110-7
[PubMed:9555893]
[WorldCat.org]
[DOI]
(P p)
P F Longchamp, C Mauël, D Karamata
Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-L-alanine amidase gene of prophage PBSX.
Microbiology (Reading): 1994, 140 ( Pt 8);1855-67
[PubMed:7921239]
[WorldCat.org]
[DOI]
(P p)