Difference between revisions of "ParA"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || forespore chromosome partitioning /<br/> negative regulation of sporulation initiation | |style="background:#ABCDEF;" align="center"|'''Function''' || forespore chromosome partitioning /<br/> negative regulation of sporulation initiation | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU40970 soj] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/Soj Soj] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/Soj Soj] | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[spo0J]]'', ''[[yyaB]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[spo0J]]'', ''[[yyaB]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU40970 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU40970 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU40970 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:soj_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:soj_context.gif]] | ||
Revision as of 14:21, 13 May 2013
- Description: ATPase, centromere-like function involved in forespore chromosome partitioning / negative regulation of sporulation initiation
| Gene name | soj |
| Synonyms | spo0JA |
| Essential | no |
| Product | negative regulator of sporulation initiation |
| Function | forespore chromosome partitioning / negative regulation of sporulation initiation |
| Gene expression levels in SubtiExpress: soj | |
| Interactions involving this protein in SubtInteract: Soj | |
| MW, pI | 27 kDa, 4.903 |
| Gene length, protein length | 759 bp, 253 aa |
| Immediate neighbours | spo0J, yyaB |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 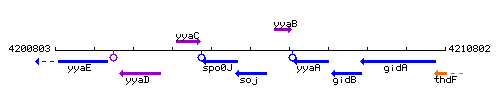
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed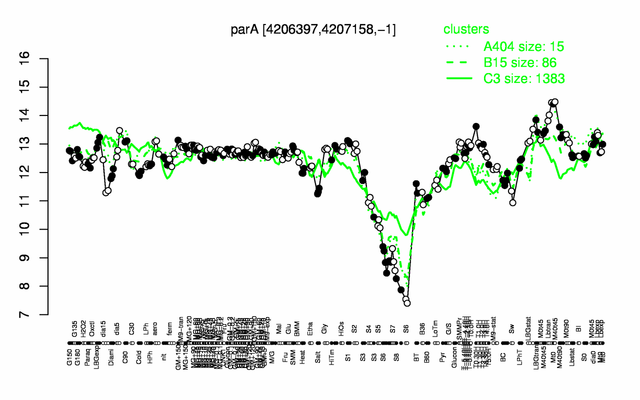
| |
Contents
Categories containing this gene/protein
DNA condensation/ segregation, sporulation/ other, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40970
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: regulates DNA replication initiation by either inhibiting or activating the DNA replication initiator protein DnaA PubMed; regulation of Spo0A expression PubMed
- Protein family: parA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P37522
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Tsutomu Katayama, Shogo Ozaki, Kenji Keyamura, Kazuyuki Fujimitsu
Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC.
Nat Rev Microbiol: 2010, 8(3);163-70
[PubMed:20157337]
[WorldCat.org]
[DOI]
(I p)
Original publications
Additional publications: PubMed
Graham Scholefield, Jeff Errington, Heath Murray
Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA.
EMBO J: 2012, 31(6);1542-55
[PubMed:22286949]
[WorldCat.org]
[DOI]
(I p)
Graham Scholefield, Rachel Whiting, Jeff Errington, Heath Murray
Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation.
Mol Microbiol: 2011, 79(4);1089-100
[PubMed:21235642]
[WorldCat.org]
[DOI]
(I p)
Henrik Strahl, Leendert W Hamoen
Membrane potential is important for bacterial cell division.
Proc Natl Acad Sci U S A: 2010, 107(27);12281-6
[PubMed:20566861]
[WorldCat.org]
[DOI]
(I p)
Stephan Gruber, Jeff Errington
Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis.
Cell: 2009, 137(4);685-96
[PubMed:19450516]
[WorldCat.org]
[DOI]
(I p)
Heath Murray, Jeff Errington
Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA.
Cell: 2008, 135(1);74-84
[PubMed:18854156]
[WorldCat.org]
[DOI]
(I p)
Gonçalo Real, Sabine Autret, Elizabeth J Harry, Jeffery Errington, Adriano O Henriques
Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis.
Mol Microbiol: 2005, 55(2);349-67
[PubMed:15659156]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Yoshitoshi Ogura, Naotake Ogasawara, Elizabeth J Harry, Shigeki Moriya
Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis.
J Bacteriol: 2003, 185(21);6316-24
[PubMed:14563866]
[WorldCat.org]
[DOI]
(P p)
Ling Juan Wu, Jeff Errington
RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis.
Mol Microbiol: 2003, 49(6);1463-75
[PubMed:12950914]
[WorldCat.org]
[DOI]
(P p)
Sabine Autret, Jeffery Errington
A role for division-site-selection protein MinD in regulation of internucleoid jumping of Soj (ParA) protein in Bacillus subtilis.
Mol Microbiol: 2003, 47(1);159-69
[PubMed:12492861]
[WorldCat.org]
[DOI]
(P p)
S Autret, R Nair, J Errington
Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein.
Mol Microbiol: 2001, 41(3);743-55
[PubMed:11532141]
[WorldCat.org]
[DOI]
(P p)
J D Quisel, A D Grossman
Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB).
J Bacteriol: 2000, 182(12);3446-51
[PubMed:10852876]
[WorldCat.org]
[DOI]
(P p)
A L Marston, J Errington
Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation.
Mol Cell: 1999, 4(5);673-82
[PubMed:10619015]
[WorldCat.org]
[DOI]
(P p)
M A Cervin, G B Spiegelman, B Raether, K Ohlsen, M Perego, J A Hoch
A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis.
Mol Microbiol: 1998, 29(1);85-95
[PubMed:9701805]
[WorldCat.org]
[DOI]
(P p)
M E Sharpe, J Errington
The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning.
Mol Microbiol: 1996, 21(3);501-9
[PubMed:8866474]
[WorldCat.org]
[DOI]
(P p)