Difference between revisions of "IlvC"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || biosynthesis of branched-chain amino acids | |style="background:#ABCDEF;" align="center"|'''Function''' || biosynthesis of branched-chain amino acids | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU28290 ilvC] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/ile_val_leu.html Ile, Leu, Val], [http://subtiwiki.uni-goettingen.de/pathways/CoA_synthesis.html Coenzyme A]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/ile_val_leu.html Ile, Leu, Val], [http://subtiwiki.uni-goettingen.de/pathways/CoA_synthesis.html Coenzyme A]''' | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[leuA]]'', ''[[ilvH]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[leuA]]'', ''[[ilvH]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU28290 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU28290 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU28290 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ilvC_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ilvC_context.gif]] | ||
Revision as of 13:30, 13 May 2013
- Description: ketol-acid reductoisomerase (2,3-dihydroxy-3-methylbutanoate, 2-acetolactate)
| Gene name | ilvC |
| Synonyms | |
| Essential | no |
| Product | ketol-acid reductoisomerase (2,3-dihydroxy-3-methylbutanoate, 2-acetolactate) |
| Function | biosynthesis of branched-chain amino acids |
| Gene expression levels in SubtiExpress: ilvC | |
| Metabolic function and regulation of this protein in SubtiPathways: Ile, Leu, Val, Coenzyme A | |
| MW, pI | 37 kDa, 5.37 |
| Gene length, protein length | 1026 bp, 342 aa |
| Immediate neighbours | leuA, ilvH |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 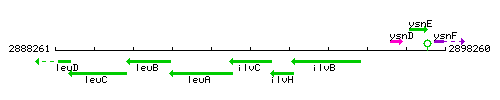
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, phosphoproteins
This gene is a member of the following regulons
CcpA regulon, CodY regulon, T-box, TnrA regulon
The gene
Basic information
- Locus tag: BSU28290
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (R)-2,3-dihydroxy-3-methylbutanoate + NADP+ = (S)-2-hydroxy-2-methyl-3-oxobutanoate + NADPH (according to Swiss-Prot)
- Protein family: ketol-acid reductoisomerase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P37253
- KEGG entry: [3]
- E.C. number: 1.1.1.86
Additional information
Expression and regulation
- Regulation: for a complete overview on the regulation of the ilv operon, see Brinsmade et al.
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Shaun R Brinsmade, Roelco J Kleijn, Uwe Sauer, Abraham L Sonenshein
Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools.
J Bacteriol: 2010, 192(24);6357-68
[PubMed:20935095]
[WorldCat.org]
[DOI]
(I p)
Boumediene Soufi, Chanchal Kumar, Florian Gnad, Matthias Mann, Ivan Mijakovic, Boris Macek
Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis.
J Proteome Res: 2010, 9(7);3638-46
[PubMed:20509597]
[WorldCat.org]
[DOI]
(I p)
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)
Shigeo Tojo, Takenori Satomura, Kanako Kumamoto, Kazutake Hirooka, Yasutaro Fujita
Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids.
J Bacteriol: 2008, 190(18);6134-47
[PubMed:18641142]
[WorldCat.org]
[DOI]
(I p)
Shigeo Tojo, Takenori Satomura, Kaori Morisaki, Ken-Ichi Yoshida, Kazutake Hirooka, Yasutaro Fujita
Negative transcriptional regulation of the ilv-leu operon for biosynthesis of branched-chain amino acids through the Bacillus subtilis global regulator TnrA.
J Bacteriol: 2004, 186(23);7971-9
[PubMed:15547269]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Susanne Hennig, Michael Hecker, Georg Homuth
Transcriptional organization and posttranscriptional regulation of the Bacillus subtilis branched-chain amino acid biosynthesis genes.
J Bacteriol: 2004, 186(8);2240-52
[PubMed:15060025]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
Holger Ludwig, Christoph Meinken, Anastasija Matin, Jörg Stülke
Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant.
J Bacteriol: 2002, 184(18);5174-8
[PubMed:12193635]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
F J Grundy, T M Henkin
Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in gram-positive bacteria.
J Mol Biol: 1994, 235(2);798-804
[PubMed:8289305]
[WorldCat.org]
[DOI]
(P p)