Difference between revisions of "RnjB"
| Line 39: | Line 39: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 111: | Line 107: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rnjB_1749418_1751085_1 rnjB] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rnjB_1749418_1751085_1 rnjB] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 143: | Line 139: | ||
=References= | =References= | ||
==Reviews== | ==Reviews== | ||
| − | '''Additional reviews:''' {{PubMed|21957024}} | + | '''Additional reviews:''' {{PubMed|23403287,21957024}} |
<big>''Lehnik-Habrink M, Lewis RJ, Mäder U, Stülke J'' </big> | <big>''Lehnik-Habrink M, Lewis RJ, Mäder U, Stülke J'' </big> | ||
<big>'''RNA degradation in ''Bacillus subtilis'': an interplay of</big> | <big>'''RNA degradation in ''Bacillus subtilis'': an interplay of</big> | ||
Revision as of 12:48, 14 February 2013
- Description: RNase J2
| Gene name | rnjB |
| Synonyms | ymfA |
| Essential | no |
| Product | RNase J2 |
| Function | RNA processing and degradation |
| Gene expression levels in SubtiExpress: rnjB | |
| Interactions involving this protein in SubtInteract: RNase J2 | |
| MW, pI | 56 kDa, 9.18 |
| Gene length, protein length | 1545 bp, 515 aa |
| Immediate neighbours | dapA, tepA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 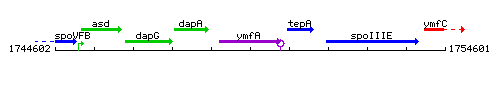
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16780
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: endoribonuclease, involved in processing of thrS mRNA
- Protein family: RNase J subfamily (according to Swiss-Prot)
- Paralogous protein(s): RnjA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- associated to the RNA degradosome PubMed
- RnjA-RnjB PubMed
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP45 (spc), GP1113 (miniTn10 spc), both available in Stülke lab
- Expression vector:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Harald Putzer, IBPC Paris, France Homepage
Your additional remarks
References
Reviews
Additional reviews: PubMed
Lehnik-Habrink M, Lewis RJ, Mäder U, Stülke J RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol Microbiol.: 2012, 84(6) 1005-1017. PubMed:22568516
Original publications
Additional publications: PubMed