Difference between revisions of "Rnr"
| Line 24: | Line 24: | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:rnr_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:rnr_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rnr_3451863_3454202_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:rnr_expression.png|500px]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | |||
<br/><br/> | <br/><br/> | ||
Revision as of 09:12, 23 April 2012
- Description: RNase R
| Gene name | rnr |
| Synonyms | yvaJ |
| Essential | no |
| Product | exoribonuclease RNase R (EC 3.1.-.-) |
| Function | nonspecific degradation of rRNA |
| MW, pI | 88 kDa, 5.703 |
| Gene length, protein length | 2337 bp, 779 aa |
| Immediate neighbours | smpB, yvaK |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 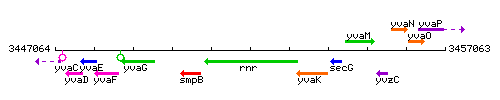
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed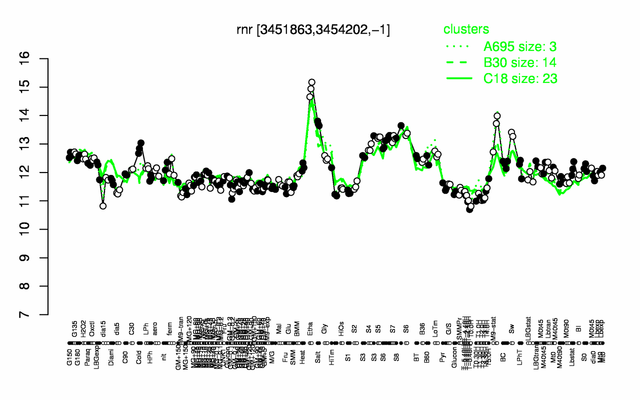
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of nucleotides, Rnases
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33610
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 3'-5'-exoribonuclease
- Protein family: ribonuclease II (RNB) family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O32231
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
David Bechhofer, Mount Sinai School, New York, USA Homepage
Your additional remarks
References
Juan Campos-Guillén, Jackeline Lizzeta Arvizu-Gómez, George H Jones, Gabriela Olmedo-Alvarez
Characterization of tRNA(Cys) processing in a conditional Bacillus subtilis CCase mutant reveals the participation of RNase R in its quality control.
Microbiology (Reading): 2010, 156(Pt 7);2102-2111
[PubMed:20360175]
[WorldCat.org]
[DOI]
(I p)
Ji-Hyun Shin, Chester W Price
The SsrA-SmpB ribosome rescue system is important for growth of Bacillus subtilis at low and high temperatures.
J Bacteriol: 2007, 189(10);3729-37
[PubMed:17369301]
[WorldCat.org]
[DOI]
(P p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Irina A Oussenko, Teppei Abe, Hiromi Ujiie, Akira Muto, David H Bechhofer
Participation of 3'-to-5' exoribonucleases in the turnover of Bacillus subtilis mRNA.
J Bacteriol: 2005, 187(8);2758-67
[PubMed:15805522]
[WorldCat.org]
[DOI]
(P p)