Difference between revisions of "YngG"
(→Expression and regulation) |
|||
| Line 99: | Line 99: | ||
* '''Operon:''' ''[[yngJ]]-[[yngI]]-[[yngH]]-[[yngHB]]-[[yngG]]-[[yngF]]-[[yngE]]'' {{PubMed|12662922}} | * '''Operon:''' ''[[yngJ]]-[[yngI]]-[[yngH]]-[[yngHB]]-[[yngG]]-[[yngF]]-[[yngE]]'' {{PubMed|12662922}} | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yngG_1952031_1952930_-1 yngG] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' [[SigE]] {{PubMed|15699190,12662922}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 09:58, 13 April 2012
- Description: 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) lyase
| Gene name | yngG |
| Synonyms | |
| Essential | no |
| Product | 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) lyase |
| Function | mother cell metabolism, leucine utilization |
| MW, pI | 32 kDa, 5.753 |
| Gene length, protein length | 897 bp, 299 aa |
| Immediate neighbours | yngF, yngH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 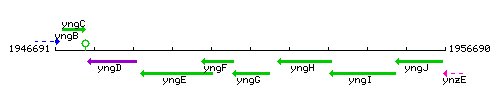
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
utilization of amino acids, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU18230
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (S)-3-hydroxy-3-methylglutaryl-CoA = acetyl-CoA + acetoacetate PubMed
- Protein family: HMG-CoA lyase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure: 1YDO
- UniProt: O34873
- KEGG entry: [3]
- E.C. number: 4.1.3.4
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Tzu-Lin Hsiao, Olga Revelles, Lifeng Chen, Uwe Sauer, Dennis Vitkup
Automatic policing of biochemical annotations using genomic correlations.
Nat Chem Biol: 2010, 6(1);34-40
[PubMed:19935659]
[WorldCat.org]
[DOI]
(I p)
Raghavendra Joshi, Brian B McSpadden Gardener
Identification and Characterization of Novel Genetic Markers Associated with Biological Control Activities in Bacillus subtilis.
Phytopathology: 2006, 96(2);145-54
[PubMed:18943917]
[WorldCat.org]
[DOI]
(P p)
Farhad Forouhar, Munif Hussain, Ramy Farid, Jordi Benach, Mariam Abashidze, William C Edstrom, Sergey M Vorobiev, Rong Xiao, Thomas B Acton, Zhuji Fu, Jung-Ja P Kim, Henry M Miziorko, Gaetano T Montelione, John F Hunt
Crystal structures of two bacterial 3-hydroxy-3-methylglutaryl-CoA lyases suggest a common catalytic mechanism among a family of TIM barrel metalloenzymes cleaving carbon-carbon bonds.
J Biol Chem: 2006, 281(11);7533-45
[PubMed:16330546]
[WorldCat.org]
[DOI]
(P p)
Leif Steil, Mónica Serrano, Adriano O Henriques, Uwe Völker
Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 2);399-420
[PubMed:15699190]
[WorldCat.org]
[DOI]
(P p)
Patrick Eichenberger, Shane T Jensen, Erin M Conlon, Christiaan van Ooij, Jessica Silvaggi, José Eduardo González-Pastor, Masaya Fujita, Sigal Ben-Yehuda, Patrick Stragier, Jun S Liu, Richard Losick
The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis.
J Mol Biol: 2003, 327(5);945-72
[PubMed:12662922]
[WorldCat.org]
[DOI]
(P p)