Difference between revisions of "SucC"
(→Extended information on the protein) |
|||
| Line 48: | Line 48: | ||
* '''Locus tag:''' BSU16090 | * '''Locus tag:''' BSU16090 | ||
| + | |||
| + | [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=sucC_1680431_1681588_1 Expression] | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
Revision as of 09:27, 25 January 2012
- Description: succinyl-CoA synthetase (beta subunit)
| Gene name | sucC |
| Synonyms | |
| Essential | no |
| Product | succinyl-CoA synthetase (beta subunit) |
| Function | TCA cycle |
| Interactions involving this protein in SubtInteract: SucC | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 41 kDa, 4.846 |
| Gene length, protein length | 1155 bp, 385 aa |
| Immediate neighbours | ylqH, sucD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 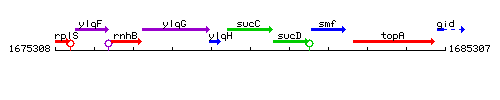
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
ATP synthesis, carbon core metabolism, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16090
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + succinate + CoA = ADP + phosphate + succinyl-CoA (according to Swiss-Prot)
- Protein family: ATP-grasp domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Reversible Michaelis-Menten FEBS Letters
- Domains:
- Modification: phosphorylation on Ser-220 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Inhibited by 2-oxoglutarate, ATP and NADH FEBS Letters
- GTP is not accept by the enzyme FEBS Letters
- Inhibited by 2-oxoglutarate, ATP and NADH FEBS Letters
Database entries
- Structure: 1JKJ (E. coli)
- UniProt: P80886
- KEGG entry: [3]
- E.C. number: 6.2.1.5
Additional information
- extensive information on the structure and enzymatic properties of succinyl-CoA synthetase can be found at Proteopedia
Expression and regulation
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Frederik M Meyer, Jan Gerwig, Elke Hammer, Christina Herzberg, Fabian M Commichau, Uwe Völker, Jörg Stülke
Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon.
Metab Eng: 2011, 13(1);18-27
[PubMed:20933603]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Ciarán Condon, Jordi Rourera, Dominique Brechemier-Baey, Harald Putzer
Ribonuclease M5 has few, if any, mRNA substrates in Bacillus subtilis.
J Bacteriol: 2002, 184(10);2845-9
[PubMed:11976317]
[WorldCat.org]
[DOI]
(P p)