Difference between revisions of "RnpA"
(→Original Publications) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' protein component of [[RNase]] P, metal-dependent 5' end maturation of precursor [[ | + | * '''Description:''' protein component of [[RNase]] P, metal-dependent 5' end maturation of precursor [[tRNA]]s <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || protein component of [[RNase]] P (substrate specificity) | |style="background:#ABCDEF;" align="center"| '''Product''' || protein component of [[RNase]] P (substrate specificity) | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || cleavage of precursor sequences <br/>from the 5' ends of pre-[[ | + | |style="background:#ABCDEF;" align="center"|'''Function''' || cleavage of precursor sequences <br/>from the 5' ends of pre-[[tRNA]]s |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 13 kDa, 10.804 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 13 kDa, 10.804 | ||
| Line 61: | Line 61: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' Endonucleolytic cleavage of RNA, removing 5'-extranucleotides from tRNA precursor (according to Swiss-Prot) | + | * '''Catalyzed reaction/ biological activity:''' Endonucleolytic cleavage of RNA, removing 5'-extranucleotides from [[tRNA]] precursor (according to Swiss-Prot) |
* '''Protein family:''' rnpA family (according to Swiss-Prot) | * '''Protein family:''' rnpA family (according to Swiss-Prot) | ||
| Line 80: | Line 80: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' [[RnpA]]-[[RpmH]] | + | * '''[[Interactions]]:''' |
| + | ** [[RnpA]]-[[RpmH]] | ||
| + | ** [[RnpA]]-[[RnpB]] {{PubMed|21622899}} | ||
* '''Localization:''' | * '''Localization:''' | ||
| Line 88: | Line 90: | ||
* '''Structure:''' | * '''Structure:''' | ||
** [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1A6F 1AF6] [http://www.ncbi.nlm.nih.gov/sites/entrez/9563955 PubMed] | ** [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1A6F 1AF6] [http://www.ncbi.nlm.nih.gov/sites/entrez/9563955 PubMed] | ||
| − | ** [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=3OKB 3OKB] (complex of RnpA with the RNA RnpB and tRNA, from ''Thermotoga maritima'') {{PubMed|21076397}} | + | ** [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=3OKB 3OKB] (complex of RnpA with the RNA [[RnpB]] and [[tRNA]], from ''Thermotoga maritima'') {{PubMed|21076397}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P25814 P25814] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P25814 P25814] | ||
Revision as of 08:29, 2 June 2011
| Gene name | rnpA |
| Synonyms | |
| Essential | yes PubMed |
| Product | protein component of RNase P (substrate specificity) |
| Function | cleavage of precursor sequences from the 5' ends of pre-tRNAs |
| MW, pI | 13 kDa, 10.804 |
| Gene length, protein length | 348 bp, 116 aa |
| Immediate neighbours | spoIIIJ, rpmH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 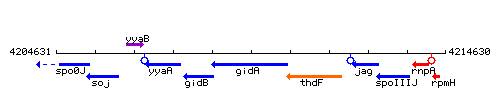
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
Rnases, translation, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU41050
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Endonucleolytic cleavage of RNA, removing 5'-extranucleotides from tRNA precursor (according to Swiss-Prot)
- Protein family: rnpA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- UniProt: P25814
- KEGG entry: [2]
- E.C. number: 3.1.26.5
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Carol Fierke, University of Michigan, Ann Arbor, USA homepage
- Roland Hartmann, Marburg University, Germany homepage
Your additional remarks
References
Reviews
Roland K Hartmann, Markus Gössringer, Bettina Späth, Susan Fischer, Anita Marchfelder
The making of tRNAs and more - RNase P and tRNase Z.
Prog Mol Biol Transl Sci: 2009, 85;319-68
[PubMed:19215776]
[WorldCat.org]
[DOI]
(P p)
J Kristin Smith, John Hsieh, Carol A Fierke
Importance of RNA-protein interactions in bacterial ribonuclease P structure and catalysis.
Biopolymers: 2007, 87(5-6);329-38
[PubMed:17868095]
[WorldCat.org]
[DOI]
(P p)
Alexei V Kazantsev, Norman R Pace
Bacterial RNase P: a new view of an ancient enzyme.
Nat Rev Microbiol: 2006, 4(10);729-40
[PubMed:16980936]
[WorldCat.org]
[DOI]
(I p)
Donald Evans, Steven M Marquez, Norman R Pace
RNase P: interface of the RNA and protein worlds.
Trends Biochem Sci: 2006, 31(6);333-41
[PubMed:16679018]
[WorldCat.org]
[DOI]
(P p)
Alfredo Torres-Larios, Kerren K Swinger, Tao Pan, Alfonso Mondragón
Structure of ribonuclease P--a universal ribozyme.
Curr Opin Struct Biol: 2006, 16(3);327-35
[PubMed:16650980]
[WorldCat.org]
[DOI]
(P p)
John Hsieh, Andy J Andrews, Carol A Fierke
Roles of protein subunits in RNA-protein complexes: lessons from ribonuclease P.
Biopolymers: 2004, 73(1);79-89
[PubMed:14691942]
[WorldCat.org]
[DOI]
(P p)
Enno Hartmann, Roland K Hartmann
The enigma of ribonuclease P evolution.
Trends Genet: 2003, 19(10);561-9
[PubMed:14550630]
[WorldCat.org]
[DOI]
(P p)
L A Kirsebom, A Vioque
RNase P from bacteria. Substrate recognition and function of the protein subunit.
Mol Biol Rep: 1995, 22(2-3);99-109
[PubMed:8901495]
[WorldCat.org]
[DOI]
(P p)
Original Publications
Additional publications: PubMed
Yu-Chu Chang, Terrence G Oas
Osmolyte-induced folding of an intrinsically disordered protein: folding mechanism in the absence of ligand.
Biochemistry: 2010, 49(25);5086-96
[PubMed:20476778]
[WorldCat.org]
[DOI]
(I p)
Kristin S Koutmou, Nathan H Zahler, Jeffrey C Kurz, Frank E Campbell, Michael E Harris, Carol A Fierke
Protein-precursor tRNA contact leads to sequence-specific recognition of 5' leaders by bacterial ribonuclease P.
J Mol Biol: 2010, 396(1);195-208
[PubMed:19932118]
[WorldCat.org]
[DOI]
(I p)
John Hsieh, Carol A Fierke
Conformational change in the Bacillus subtilis RNase P holoenzyme--pre-tRNA complex enhances substrate affinity and limits cleavage rate.
RNA: 2009, 15(8);1565-77
[PubMed:19549719]
[WorldCat.org]
[DOI]
(I p)
Markus Gösringer, Roland K Hartmann
Function of heterologous and truncated RNase P proteins in Bacillus subtilis.
Mol Microbiol: 2007, 66(3);801-13
[PubMed:17919279]
[WorldCat.org]
[DOI]
(P p)
Barbara Wegscheid, Roland K Hartmann
In vivo and in vitro investigation of bacterial type B RNase P interaction with tRNA 3'-CCA.
Nucleic Acids Res: 2007, 35(6);2060-73
[PubMed:17355991]
[WorldCat.org]
[DOI]
(I p)
Somashekarappa Niranjanakumari, Jeremy J Day-Storms, Mahiuddin Ahmed, John Hsieh, Nathan H Zahler, Ronald A Venters, Carol A Fierke
Probing the architecture of the B. subtilis RNase P holoenzyme active site by cross-linking and affinity cleavage.
RNA: 2007, 13(4);521-35
[PubMed:17299131]
[WorldCat.org]
[DOI]
(P p)
Markus Gössringer, Rosel Kretschmer-Kazemi Far, Roland K Hartmann
Analysis of RNase P protein (rnpA) expression in Bacillus subtilis utilizing strains with suppressible rnpA expression.
J Bacteriol: 2006, 188(19);6816-23
[PubMed:16980484]
[WorldCat.org]
[DOI]
(P p)
Christopher H Henkels, Terrence G Oas
Thermodynamic characterization of the osmolyte- and ligand-folded states of Bacillus subtilis ribonuclease P protein.
Biochemistry: 2005, 44(39);13014-26
[PubMed:16185070]
[WorldCat.org]
[DOI]
(P p)
Christoph Rox, Ralph Feltens, Thomas Pfeiffer, Roland K Hartmann
Potential contact sites between the protein and RNA subunit in the Bacillus subtilis RNase P holoenzyme.
J Mol Biol: 2002, 315(4);551-60
[PubMed:11812129]
[WorldCat.org]
[DOI]
(P p)
A Hansen, T Pfeiffer, T Zuleeg, S Limmer, J Ciesiolka, R Feltens, R K Hartmann
Exploring the minimal substrate requirements for trans-cleavage by RNase P holoenzymes from Escherichia coli and Bacillus subtilis.
Mol Microbiol: 2001, 41(1);131-43
[PubMed:11454206]
[WorldCat.org]
[DOI]
(P p)
C H Henkels, J C Kurz, C A Fierke, T G Oas
Linked folding and anion binding of the Bacillus subtilis ribonuclease P protein.
Biochemistry: 2001, 40(9);2777-89
[PubMed:11258888]
[WorldCat.org]
[DOI]
(P p)
J M Warnecke, R Held, S Busch, R K Hartmann
Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis.
J Mol Biol: 1999, 290(2);433-45
[PubMed:10390342]
[WorldCat.org]
[DOI]
(P p)
T Stams, S Niranjanakumari, C A Fierke, D W Christianson
Ribonuclease P protein structure: evolutionary origins in the translational apparatus.
Science: 1998, 280(5364);752-5
[PubMed:9563955]
[WorldCat.org]
[DOI]
(P p)