Difference between revisions of "YbfO"
(→Expression and regulation) |
|||
| Line 98: | Line 98: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''ybfO'' (according to [http://dbtbs.hgc.jp/COG/prom/ybfO.html DBTBS]) | + | * '''Operon:''' |
| + | ** ''ybfO'' (according to [http://dbtbs.hgc.jp/COG/prom/ybfO.html DBTBS]) | ||
| + | ** ''[[ybfO]]-[[ybfP]]'' {{PubMed|20817675}} | ||
| + | ** ''[[ybfO]]-[[ybfP]]-[[ybfQ]]'' {{PubMed|12207695}} | ||
* '''[[Sigma factor]]:''' [[SigW]] {{PubMed|11866510}} | * '''[[Sigma factor]]:''' [[SigW]] {{PubMed|11866510}} | ||
| Line 109: | Line 112: | ||
** [[AbrB]]: transcription repression {{PubMed|18840696,20817675}} | ** [[AbrB]]: transcription repression {{PubMed|18840696,20817675}} | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 17:42, 17 May 2011
- Description: similar to erythromycin esterase
| Gene name | ybfO |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| MW, pI | 51 kDa, 6.678 |
| Gene length, protein length | 1338 bp, 446 aa |
| Immediate neighbours | ybfN, ybfP |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 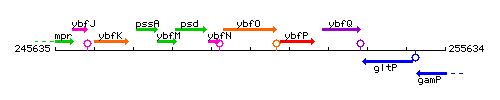
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
cell envelope stress proteins (controlled by SigM, W, X, Y), resistance against toxins/ antibiotics
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU02310
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: extracellular (signal peptide) PubMed
Database entries
- Structure:
- UniProt: O31455
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Allison V Banse, Arnaud Chastanet, Lilah Rahn-Lee, Errett C Hobbs, Richard Losick
Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2008, 105(40);15547-52
[PubMed:18840696]
[WorldCat.org]
[DOI]
(I p)
Min Cao, Phil A Kobel, Maud M Morshedi, Ming Fang Winston Wu, Chris Paddon, John D Helmann
Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches.
J Mol Biol: 2002, 316(3);443-57
[PubMed:11866510]
[WorldCat.org]
[DOI]
(P p)