Difference between revisions of "MtrB"
(→Database entries) |
|||
| Line 94: | Line 94: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId= | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=3AQD 3AQD], [http://www.rcsb.org/pdb/explore.do?structureId=1WAP 1WAP] (complex with L-tryptophan), [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2ZP9 2ZP9] (complex with [[RtpA]]) |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P19466 P19466] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P19466 P19466] | ||
Revision as of 16:38, 10 January 2011
- Description: tryptophan operon RNA-binding attenuation protein (TRAP), controls the RNA switch in front of genes involved in biosynthesis and acquisition of tryptophan
| Gene name | mtrB |
| Synonyms | |
| Essential | no |
| Product | tryptophan operon RNA-binding attenuation protein (TRAP) |
| Function | regulation of tryptophan biosynthesis (and translation) attenuation in the trp operon; repression of the folate operon |
| Metabolic function and regulation of this protein in SubtiPathways: Phe, Tyr, Trp, Folate | |
| MW, pI | 8 kDa, 7.333 |
| Gene length, protein length | 225 bp, 75 aa |
| Immediate neighbours | hepS, folE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 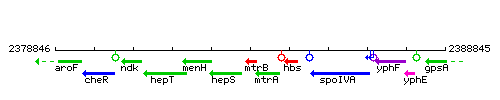
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, transcription factors and their control, RNA binding regulators
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22770
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: prpD family (according to Swiss-Prot)
- Paralogous protein(s):
Genes controlled by MtrB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- UniProt: P19466
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications
Kristine D Potter, Natalie M Merlino, Timothy Jacobs, Paul Gollnick
TRAP binding to the Bacillus subtilis trp leader region RNA causes efficient transcription termination at a weak intrinsic terminator.
Nucleic Acids Res: 2011, 39(6);2092-102
[PubMed:21097886]
[WorldCat.org]
[DOI]
(I p)
Adam P McGraw, Ali Mokdad, François Major, Philip C Bevilacqua, Paul Babitzke
Molecular basis of TRAP-5'SL RNA interaction in the Bacillus subtilis trp operon transcription attenuation mechanism.
RNA: 2009, 15(1);55-66
[PubMed:19033375]
[WorldCat.org]
[DOI]
(I p)
Yanling Chen, Paul Gollnick
Alanine scanning mutagenesis of anti-TRAP (AT) reveals residues involved in binding to TRAP.
J Mol Biol: 2008, 377(5);1529-43
[PubMed:18334255]
[WorldCat.org]
[DOI]
(I p)
Adam P McGraw, Philip C Bevilacqua, Paul Babitzke
TRAP-5' stem loop interaction increases the efficiency of transcription termination in the Bacillus subtilis trpEDCFBA operon leader region.
RNA: 2007, 13(11);2020-33
[PubMed:17881743]
[WorldCat.org]
[DOI]
(P p)
Maria V Barbolina, Roman Kristoforov, Amanda Manfredo, Yanling Chen, Paul Gollnick
The rate of TRAP binding to RNA is crucial for transcription attenuation control of the B. subtilis trp operon.
J Mol Biol: 2007, 370(5);925-38
[PubMed:17555767]
[WorldCat.org]
[DOI]
(P p)
Helen Yakhnin, Alexander V Yakhnin, Paul Babitzke
Translation control of trpG from transcripts originating from the folate operon promoter of Bacillus subtilis is influenced by translation-mediated displacement of bound TRAP, while translation control of transcripts originating from a newly identified trpG promoter is not.
J Bacteriol: 2007, 189(3);872-9
[PubMed:17114263]
[WorldCat.org]
[DOI]
(P p)
Alexander V Yakhnin, Helen Yakhnin, Paul Babitzke
RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction.
Mol Cell: 2006, 24(4);547-57
[PubMed:17114058]
[WorldCat.org]
[DOI]
(P p)
Helen Yakhnin, Alexander V Yakhnin, Paul Babitzke
The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding.
Mol Microbiol: 2006, 61(5);1252-66
[PubMed:16879415]
[WorldCat.org]
[DOI]
(P p)
Craig A McElroy, Amanda Manfredo, Paul Gollnick, Mark P Foster
Thermodynamics of tryptophan-mediated activation of the trp RNA-binding attenuation protein.
Biochemistry: 2006, 45(25);7844-53
[PubMed:16784236]
[WorldCat.org]
[DOI]
(P p)
Vandana Payal, Paul Gollnick
Substitutions of Thr30 provide mechanistic insight into tryptophan-mediated activation of TRAP binding to RNA.
Nucleic Acids Res: 2006, 34(10);2933-42
[PubMed:16738132]
[WorldCat.org]
[DOI]
(I e)
Wen-Jen Yang, Charles Yanofsky
Effects of tryptophan starvation on levels of the trp RNA-binding attenuation protein (TRAP) and anti-TRAP regulatory protein and their influence on trp operon expression in Bacillus subtilis.
J Bacteriol: 2005, 187(6);1884-91
[PubMed:15743934]
[WorldCat.org]
[DOI]
(P p)
Maria V Barbolina, Xiufeng Li, Paul Gollnick
Bacillus subtilis TRAP binds to its RNA target by a 5' to 3' directional mechanism.
J Mol Biol: 2005, 345(4);667-79
[PubMed:15588817]
[WorldCat.org]
[DOI]
(P p)
Barbara C McCabe, Paul Gollnick
Cellular levels of trp RNA-binding attenuation protein in Bacillus subtilis.
J Bacteriol: 2004, 186(15);5157-9
[PubMed:15262953]
[WorldCat.org]
[DOI]
(P p)
Doug Snyder, Jeffrey Lary, Yanling Chen, Paul Gollnick, James L Cole
Interaction of the trp RNA-binding attenuation protein (TRAP) with anti-TRAP.
J Mol Biol: 2004, 338(4);669-82
[PubMed:15099736]
[WorldCat.org]
[DOI]
(P p)
Nicholas H Hopcroft, Amanda Manfredo, Alice L Wendt, Andrzej M Brzozowski, Paul Gollnick, Alfred A Antson
The interaction of RNA with TRAP: the role of triplet repeats and separating spacer nucleotides.
J Mol Biol: 2004, 338(1);43-53
[PubMed:15050822]
[WorldCat.org]
[DOI]
(P p)
Gintaras Deikus, Paul Babitzke, David H Bechhofer
Recycling of a regulatory protein by degradation of the RNA to which it binds.
Proc Natl Acad Sci U S A: 2004, 101(9);2747-51
[PubMed:14976255]
[WorldCat.org]
[DOI]
(P p)
Helen Yakhnin, Hong Zhang, Alexander V Yakhnin, Paul Babitzke
The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding.
J Bacteriol: 2004, 186(2);278-86
[PubMed:14702295]
[WorldCat.org]
[DOI]
(P p)
Pan T X Li, Paul Gollnick
Characterization of a trp RNA-binding attenuation protein (TRAP) mutant with tryptophan independent RNA binding activity.
J Mol Biol: 2004, 335(3);707-22
[PubMed:14687568]
[WorldCat.org]
[DOI]
(P p)
Janell E Schaak, Helen Yakhnin, Philip C Bevilacqua, Paul Babitzke
A Mg2+-dependent RNA tertiary structure forms in the Bacillus subtilis trp operon leader transcript and appears to interfere with trpE translation control by inhibiting TRAP binding.
J Mol Biol: 2003, 332(3);555-74
[PubMed:12963367]
[WorldCat.org]
[DOI]
(P p)
Pan T X Li, Paul Gollnick
Using hetero-11-mers composed of wild type and mutant subunits to study tryptophan binding to TRAP and its role in activating RNA binding.
J Biol Chem: 2002, 277(38);35567-73
[PubMed:12133840]
[WorldCat.org]
[DOI]
(P p)
Nicholas H Hopcroft, Alice L Wendt, Paul Gollnick, Alfred A Antson
Specificity of TRAP-RNA interactions: crystal structures of two complexes with different RNA sequences.
Acta Crystallogr D Biol Crystallogr: 2002, 58(Pt 4);615-21
[PubMed:11914485]
[WorldCat.org]
[DOI]
(P p)
Pan T X Li, David J Scott, Paul Gollnick
Creating hetero-11-mers composed of wild-type and mutant subunits to study RNA binding to TRAP.
J Biol Chem: 2002, 277(14);11838-44
[PubMed:11805104]
[WorldCat.org]
[DOI]
(P p)
Angela Valbuzzi, Paul Gollnick, Paul Babitzke, Charles Yanofsky
The anti-trp RNA-binding attenuation protein (Anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein.
J Biol Chem: 2002, 277(12);10608-13
[PubMed:11786553]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, P Gollnick
Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the trp RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA structure.
J Bacteriol: 2001, 183(20);5795-802
[PubMed:11566976]
[WorldCat.org]
[DOI]
(P p)
A Valbuzzi, C Yanofsky
Inhibition of the B. subtilis regulatory protein TRAP by the TRAP-inhibitory protein, AT.
Science: 2001, 293(5537);2057-9
[PubMed:11557884]
[WorldCat.org]
[DOI]
(P p)
M B Elliott, P A Gottlieb, P Gollnick
The mechanism of RNA binding to TRAP: initiation and cooperative interactions.
RNA: 2001, 7(1);85-93
[PubMed:11214184]
[WorldCat.org]
[DOI]
(P p)
J P Sarsero, E Merino, C Yanofsky
A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport.
J Bacteriol: 2000, 182(8);2329-31
[PubMed:10735881]
[WorldCat.org]
[DOI]
(P p)
H Du, A V Yakhnin, S Dharmaraj, P Babitzke
trp RNA-binding attenuation protein-5' stem-loop RNA interaction is required for proper transcription attenuation control of the Bacillus subtilis trpEDCFBA operon.
J Bacteriol: 2000, 182(7);1819-27
[PubMed:10714985]
[WorldCat.org]
[DOI]
(P p)
A V Yakhnin, J J Trimble, C R Chiaro, P Babitzke
Effects of mutations in the L-tryptophan binding pocket of the Trp RNA-binding attenuation protein of Bacillus subtilis.
J Biol Chem: 2000, 275(6);4519-24
[PubMed:10660627]
[WorldCat.org]
[DOI]
(P p)
A A Antson, E J Dodson, G Dodson, R B Greaves, X Chen, P Gollnick
Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA.
Nature: 1999, 401(6750);235-42
[PubMed:10499579]
[WorldCat.org]
[DOI]
(P p)
X p Chen, A A Antson, M Yang, P Li, C Baumann, E J Dodson, G G Dodson, P Gollnick
Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus.
J Mol Biol: 1999, 289(4);1003-16
[PubMed:10369778]
[WorldCat.org]
[DOI]
(P p)
S Xirasagar, M B Elliott, W Bartolini, P Gollnick, P A Gottlieb
RNA structure inhibits the TRAP (trp RNA-binding attenuation protein)-RNA interaction.
J Biol Chem: 1998, 273(42);27146-53
[PubMed:9765233]
[WorldCat.org]
[DOI]
(P p)
C Baumann, S Xirasagar, P Gollnick
The trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis binds to unstacked trp leader RNA.
J Biol Chem: 1997, 272(32);19863-9
[PubMed:9242649]
[WorldCat.org]
[DOI]
(P p)
M Yang, X p Chen, K Militello, R Hoffman, B Fernandez, C Baumann, P Gollnick
Alanine-scanning mutagenesis of Bacillus subtilis trp RNA-binding attenuation protein (TRAP) reveals residues involved in tryptophan binding and RNA binding.
J Mol Biol: 1997, 270(5);696-710
[PubMed:9245598]
[WorldCat.org]
[DOI]
(P p)
A I Lee, J P Sarsero, C Yanofsky
A temperature-sensitive trpS mutation interferes with trp RNA-binding attenuation protein (TRAP) regulation of trp gene expression in Bacillus subtilis.
J Bacteriol: 1996, 178(22);6518-24
[PubMed:8932308]
[WorldCat.org]
[DOI]
(P p)
C Baumann, J Otridge, P Gollnick
Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA.
J Biol Chem: 1996, 271(21);12269-74
[PubMed:8647825]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, D G Bear, C Yanofsky
TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides.
Proc Natl Acad Sci U S A: 1995, 92(17);7916-20
[PubMed:7544009]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, C Yanofsky
Structural features of L-tryptophan required for activation of TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis.
J Biol Chem: 1995, 270(21);12452-6
[PubMed:7759487]
[WorldCat.org]
[DOI]
(P p)
A A Antson, J Otridge, A M Brzozowski, E J Dodson, G G Dodson, K S Wilson, T M Smith, M Yang, T Kurecki, P Gollnick
The structure of trp RNA-binding attenuation protein.
Nature: 1995, 374(6524);693-700
[PubMed:7715723]
[WorldCat.org]
[DOI]
(P p)
P Gollnick, C Baumann, M Yang, J Otridge, A Antson
Interaction of the 11-subunit trp RNA-binding attenuation protein (TRAP) with its RNA target.
Nucleic Acids Symp Ser: 1995, (33);43-5
[PubMed:8643393]
[WorldCat.org]
(P p)
A A Antson, A M Brzozowski, E J Dodson, Z Dauter, K S Wilson, T Kurecki, J Otridge, P Gollnick
11-fold symmetry of the trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis determined by X-ray analysis.
J Mol Biol: 1994, 244(1);1-5
[PubMed:7525975]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, J T Stults, S J Shire, C Yanofsky
TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts.
J Biol Chem: 1994, 269(24);16597-604
[PubMed:7515880]
[WorldCat.org]
(P p)
P Babitzke, C Yanofsky
Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein.
Proc Natl Acad Sci U S A: 1993, 90(1);133-7
[PubMed:7678334]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, P Gollnick, C Yanofsky
The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of tryptophan biosynthesis.
J Bacteriol: 1992, 174(7);2059-64
[PubMed:1551827]
[WorldCat.org]
[DOI]
(P p)
P Gollnick, S Ishino, M I Kuroda, D J Henner, C Yanofsky
The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis.
Proc Natl Acad Sci U S A: 1990, 87(22);8726-30
[PubMed:2123343]
[WorldCat.org]
[DOI]
(P p)