Difference between revisions of "YwlF"
(→Expression and regulation) |
Raphael2215 (talk | contribs) (→Database entries) |
||
| Line 81: | Line 81: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.pdb.org/pdb/explore/explore.do?structureId=3HEE 3HEE] (from ''Clostridium thermocellum'', 55% identity, 72% similarity) |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P39156 P39156] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P39156 P39156] | ||
Revision as of 10:52, 21 July 2010
- Description: ribose-5-phosphate isomerase
| Gene name | ywlF |
| Synonyms | ipc-32d |
| Essential | no |
| Product | ribose-5-phosphate isomerase |
| Function | pentose phosphate pathway |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 16 kDa, 5.416 |
| Gene length, protein length | 447 bp, 149 aa |
| Immediate neighbours | ywlG, ywlE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 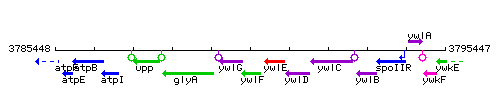
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU36920
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: lacAB/rpiB family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure: 3HEE (from Clostridium thermocellum, 55% identity, 72% similarity)
- UniProt: P39156
- KEGG entry: [3]
- E.C. number: 5.3.1.6
Additional information
The enzyme has probably more than one active site, but with no cooperativity described PubMed
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Shuobo Shi, Tao Chen, Zhigang Zhang, Xun Chen, Xueming Zhao
Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production.
Metab Eng: 2009, 11(4-5);243-52
[PubMed:19446032]
[WorldCat.org]
[DOI]
(I p)
Ken-ichi Yoshida, Hirotake Yamaguchi, Masaki Kinehara, Yo-hei Ohki, Yoshiko Nakaura, Yasutaro Fujita
Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box.
Mol Microbiol: 2003, 49(1);157-65
[PubMed:12823818]
[WorldCat.org]
[DOI]
(P p)
R D MacElroy, C R Middaugh
Bacterial ribosephosphate isomerase.
Methods Enzymol: 1982, 89 Pt D;571-9
[PubMed:7144591]
[WorldCat.org]
[DOI]
(P p)
C R Middaugh, R D MacElroy
The effect of temperature on ribose-5-phosphate isomerase from a mesophile, Thiobacillus thioparus, and a thermophile, Bacillus caldolyticus.
J Biochem: 1976, 79(6);1331-44
[PubMed:956158]
[WorldCat.org]
[DOI]
(P p)